Fusarium Photoreceptors
- PMID: 36983487
- PMCID: PMC10056346
- DOI: 10.3390/jof9030319
Fusarium Photoreceptors
Abstract
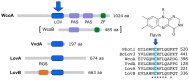
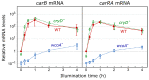
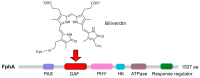
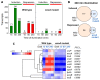
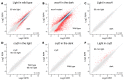
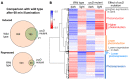
Light is an important modulating signal in fungi. Fusarium species stand out as research models for their phytopathogenic activity and their complex secondary metabolism. This includes the synthesis of carotenoids, whose induction by light is their best known photoregulated process. In these fungi, light also affects other metabolic pathways and developmental stages, such as the formation of conidia. Photoreceptor proteins are essential elements in signal transduction from light. Fusarium genomes contain genes for at least ten photoreceptors: four flavoproteins, one photolyase, two cryptochromes, two rhodopsins, and one phytochrome. Mutations in five of these genes provide information about their functions in light regulation, in which the flavoprotein WcoA, belonging to the White Collar (WC) family, plays a predominant role. Global transcriptomic techniques have opened new perspectives for the study of photoreceptor functions and have recently been used in Fusarium fujikuroi on a WC protein and a cryptochrome from the DASH family. The data showed that the WC protein participates in the transcriptional control of most of the photoregulated genes, as well as of many genes not regulated by light, while the DASH cryptochrome potentially plays a supporting role in the photoinduction of many genes.
Keywords: RNA-seq analyses; White Collar; cryptochrome; flavoprotein; light detection; phytochrome; rhodopsin.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; in the decision to publish the results.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

