Data Independent Acquisition Reveals In-Depth Serum Proteome Changes in Canine Leishmaniosis
- PMID: 36984805
- PMCID: PMC10059658
- DOI: 10.3390/metabo13030365
Data Independent Acquisition Reveals In-Depth Serum Proteome Changes in Canine Leishmaniosis
Abstract
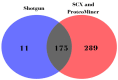
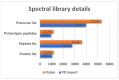
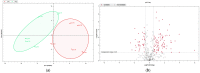
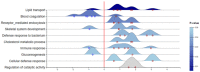
Comprehensive profiling of serum proteome provides valuable clues of health status and pathophysiological processes, making it the main strategy in biomarker discovery. However, the high dynamic range significantly decreases the number of detectable proteins, obstructing the insights into the underlying biological processes. To circumvent various serum enrichment methods, obtain high-quality proteome wide information using the next-generation proteomic, and study host response in canine leishmaniosis, we applied data-independent acquisition mass spectrometry (DIA-MS) for deep proteomic profiling of clinical samples. The non-depleted serum samples of healthy and naturally Leishmania-infected dogs were analyzed using the label-free 60-min gradient sequential window acquisition of all theoretical mass spectra (SWATH-MS) method. As a result, we identified 554 proteins, 140 of which differed significantly in abundance. Those were included in lipid metabolism, hematological abnormalities, immune response, and oxidative stress, providing valuable information about the complex molecular basis of the clinical and pathological landscape in canine leishmaniosis. Our results show that DIA-MS is a method of choice for understanding complex pathophysiological processes in serum and serum biomarker development.
Keywords: biomarker discovery; canine; data independent acquisition; leishmaniosis; mass spectrometry; proteomics; serum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Paul J., Veenstra T.D. Separation of Serum and Plasma Proteins for In-Depth Proteomic Analysis. Separations. 2022;9:89. doi: 10.3390/separations9040089. - DOI
-
- Košiček M., Gudelj I., Horvatić A., Jović T., Vučković F., Lauc G., Hećimović S., Kosicek M., Gudelj I., Horvatic A., et al. N-glycome of the lysosomal glycocalyx is altered in Niemann-Pick Type C disease model cells. Mol. Cell. Proteom. 2018;17:631–642. doi: 10.1074/mcp.RA117.000129. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

