Comparative Analysis of the Symbiotic Microbiota in the Chinese Mitten Crab (Eriocheir sinensis): Microbial Structure, Co-Occurrence Patterns, and Predictive Functions
- PMID: 36985118
- PMCID: PMC10053967
- DOI: 10.3390/microorganisms11030544
Comparative Analysis of the Symbiotic Microbiota in the Chinese Mitten Crab (Eriocheir sinensis): Microbial Structure, Co-Occurrence Patterns, and Predictive Functions
Abstract
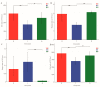
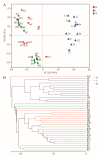
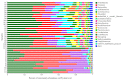
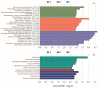
Symbiotic microorganisms in the digestive and circulatory systems are found in various crustaceans, and their essential roles in crustacean health, nutrition, and disease have attracted considerable interest. Although the intestinal microbiota of the Chinese mitten crab (Eriocheir sinensis) has been extensively studied, information on the symbiotic microbiota at various sites of this aquatic economic species, particularly the hepatopancreas and hemolymph, is lacking. This study aimed to comprehensively characterize the hemolymph, hepatopancreas, and intestinal microbiota of Chinese mitten crab through the high-throughput sequencing of the 16S rRNA gene. Results showed no significant difference in microbial diversity between the hemolymph and hepatopancreas (Welch t-test; p > 0.05), but their microbial diversity was significantly higher than that in the intestine (p < 0.05). Distinct differences were found in the structure, composition, and predicted function of the symbiotic microbiota at these sites. At the phylum level, the hemolymph and hepatopancreas microbiota were dominated by Proteobacteria, Firmicutes, and Acidobacteriota, followed by Bacteroidota and Actinobacteriota, whereas the gut microbiota was mainly composed of Firmicutes, Proteobacteria, and Bacteroidota. At the genus level, Candidatus Hepatoplasma, Shewanella, and Aeromonas were dominant in the hepatopancreas; Candidatus Bacilloplasma, Roseimarinus, and Vibrio were dominant in the intestine; Enterobacter, norank_Vicinamibacterales, and Pseudomonas were relatively high-abundance genera in the hemolymph. The composition and abundance of symbiotic microbiota in the hemolymph and hepatopancreas were extremely similar (p > 0.05), and no significant difference in functional prediction was found (p > 0.05). Comparing the hemolymph in the intestine and hepatopancreas, the hemolymph had lower variation in bacterial composition among individuals, having a more uniform abundance of major bacterial taxa, a smaller coefficient of variation, and the highest proportion of shared genera. Network complexity varied greatly among the three sites. The hepatopancreas microbiota was the most complex, followed by the hemolymph microbiota, and the intestinal microbiota had the simplest network. This study revealed the taxonomic and functional characteristics of the hemolymph, hepatopancreas, and gut microbiota in Chinese mitten crab. The results expanded our understanding of the symbiotic microbiota in crustaceans, providing potential indicators for assessing the health status of Chinese mitten crab.
Keywords: Chinese mitten crab; hemolymph; hepatopancreas; high-throughput sequencing; intestine; symbiotic microbiota.
Conflict of interest statement
The authors declare no conflict to interest.
Figures


Similar articles
-
Changes in Hemolymph Microbiota of Chinese Mitten Crab (Eriocheir sinensis) in Response to Aeromonas hydrophila or Staphylococcus aureus Infection.Animals (Basel). 2023 Sep 29;13(19):3058. doi: 10.3390/ani13193058. Animals (Basel). 2023. PMID: 37835665 Free PMC article.
-
Seasonal dynamics of intestinal microbiota in juvenile Chinese mitten crab (Eriocheir sinensis) in the Yangtze Estuary.Front Cell Infect Microbiol. 2024 Jul 4;14:1436547. doi: 10.3389/fcimb.2024.1436547. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39027136 Free PMC article.
-
Effects of Dietary L-TRP on Immunity, Antioxidant Capacity and Intestinal Microbiota of the Chinese Mitten Crab (Eriocheir Sinensis) in Pond Culture.Metabolites. 2022 Dec 20;13(1):1. doi: 10.3390/metabo13010001. Metabolites. 2022. PMID: 36676926 Free PMC article.
-
16S rRNA gene sequencing analysis reveals an imbalance in the intestinal flora of Eriocheir sinensis with hepatopancreatic necrosis disease.Comp Biochem Physiol Part D Genomics Proteomics. 2022 Jun;42:100988. doi: 10.1016/j.cbd.2022.100988. Epub 2022 Apr 9. Comp Biochem Physiol Part D Genomics Proteomics. 2022. PMID: 35468457
-
Status and prospects of product processing and sustainable utilization of Chinese mitten crab (Eriocheir sinensis).Heliyon. 2024 Jun 13;10(12):e32922. doi: 10.1016/j.heliyon.2024.e32922. eCollection 2024 Jun 30. Heliyon. 2024. PMID: 38994063 Free PMC article. Review.
Cited by
-
Changes in Hemolymph Microbiota of Chinese Mitten Crab (Eriocheir sinensis) in Response to Aeromonas hydrophila or Staphylococcus aureus Infection.Animals (Basel). 2023 Sep 29;13(19):3058. doi: 10.3390/ani13193058. Animals (Basel). 2023. PMID: 37835665 Free PMC article.
-
The shared microbiome in mud crab (Scylla paramamosain) of Sanmen Bay, China: core gut microbiome.Front Microbiol. 2023 Sep 1;14:1243334. doi: 10.3389/fmicb.2023.1243334. eCollection 2023. Front Microbiol. 2023. PMID: 37727291 Free PMC article.
References
-
- McFall-Ngai M., Hadfield M.G., Bosch T.C.G., Carey H.V., Domazet-Loso T., Douglas A.E., Dubilier N., Eberl G., Fukami T., Gilbert S.F., et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. - DOI - PMC - PubMed
-
- Bikel S., Valdez-Lara A., Cornejo-Granados F., Rico K., Canizales-Quinteros S., Soberon X., Del Pozo-Yauner L., Ochoa-Leyva A. Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: Towards a systems-level understanding of human microbiome. Comput. Struct. Biotechnol. J. 2015;13:390–401. doi: 10.1016/j.csbj.2015.06.001. - DOI - PMC - PubMed
-
- Verner-Jeffreys D.W., Shields R.J., Bricknell I.R., Birkbeck T.H. Changes in the gut-associated microflora during the development of Atlantic halibut (Hippoglossus hippoglossus L.) larvae in three British hatcheries. Aquaculture. 2003;219:21–42. doi: 10.1016/S0044-8486(02)00348-4. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

