Avian Malaria Parasites Modulate Gut Microbiome Assembly in Canaries
- PMID: 36985137
- PMCID: PMC10056159
- DOI: 10.3390/microorganisms11030563
Avian Malaria Parasites Modulate Gut Microbiome Assembly in Canaries
Abstract
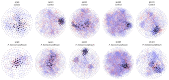
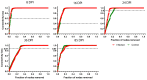
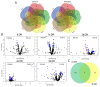
Rodent and human malaria parasites cause dysbiosis in the host gut microbiome, but whether Plasmodium species affecting birds cause dysbiosis in their hosts is currently unknown. Here we used a model of avian malaria infection to test whether parasite infection modulates the bird microbiome. To this aim, bird fecal microbiomes were characterized at different time points after infection of canaries with the avian malaria parasite Plasmodium homocircumflexum. Avian malaria caused no significant changes in the alpha and beta diversity of the microbiome in infected birds. In contrast, we discovered changes in the composition and abundance of several taxa. Co-occurrence networks were used to characterize the assembly of the microbiome and trajectories of microbiome structural states progression were found to be different between infected and uninfected birds. Prediction of functional profiles in bacterial communities using PICRUSt2 showed infection by P. homocircumflexum to be associated with the presence of specific degradation and biosynthesis metabolic pathways, which were not found in healthy birds. Some of the metabolic pathways with decreased abundance in the infected group had significant increase in the later stage of infection. The results showed that avian malaria parasites affect bacterial community assembly in the host gut microbiome. Microbiome modulation by malaria parasites could have deleterious consequences for the host bird. Knowing the intricacies of bird-malaria-microbiota interactions may prove helpful in determining key microbial players and informing interventions to improve animal health.
Keywords: avian malaria; microbiome; parasite-microbiota interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Grond K., Sandercock B.K., Jumpponen A., Zeglin L.H. The Avian Gut Microbiota: Community, Physiology and Function in Wild Birds. J. Avian Biol. 2018;49:e01788. doi: 10.1111/jav.01788. - DOI
-
- Grond K., Santo Domingo J.W., Lanctot R.B., Jumpponen A., Bentzen R.L., Boldenow M.L., Brown S.C., Casler B., Cunningham J.A., Doll A.C., et al. Composition and Drivers of Gut Microbial Communities in Arctic-Breeding Shorebirds. Front. Microbiol. 2019;10:2258. doi: 10.3389/fmicb.2019.02258. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

