Characterization of a Lytic Bacteriophage and Demonstration of Its Combined Lytic Effect with a K2 Depolymerase on the Hypervirulent Klebsiella pneumoniae Strain 52145
- PMID: 36985241
- PMCID: PMC10051899
- DOI: 10.3390/microorganisms11030669
Characterization of a Lytic Bacteriophage and Demonstration of Its Combined Lytic Effect with a K2 Depolymerase on the Hypervirulent Klebsiella pneumoniae Strain 52145
Abstract
Klebsiella pneumoniae is a nosocomial pathogen. Among its virulence factors is the capsule with a prominent role in defense and biofilm formation. Bacteriophages (phages) can evoke the lysis of bacterial cells. Due to the mode of action of their polysaccharide depolymerase enzymes, phages are typically specific for one bacterial strain and its capsule type. In this study, we characterized a bacteriophage against the capsule-defective mutant of the nosocomial K. pneumoniae 52145 strain, which lacks K2 capsule. The phage showed a relatively narrow host range but evoked lysis on a few strains with capsular serotypes K33, K21, and K24. Phylogenetic analysis showed that the newly isolated Klebsiella phage 731 belongs to the Webervirus genus in the Drexlerviridae family; it has a 31.084 MDa double-stranded, linear DNA with a length of 50,306 base pairs and a G + C content of 50.9%. Out of the 79 open reading frames (ORFs), we performed the identification of orf22, coding for a trimeric tail fiber protein with putative capsule depolymerase activity, along with the mapping of other putative depolymerases of phage 731 and homologous phages. Efficacy of a previously described recombinant K2 depolymerase (B1dep) was tested by co-spotting phage 731 on K. pneumoniae strains, and it was demonstrated that the B1dep-phage 731 combination allows the lysis of the wild type 52145 strain, originally resistant to the phage 731. With phage 731, we showed that B1dep is a promising candidate for use as a possible antimicrobial agent, as it renders the virulent strain defenseless against other phages. Phage 731 alone is also important due to its efficacy on K. pneumoniae strains possessing epidemiologically important serotypes.
Keywords: Klebsiella phage; Klebsiella pneumoniae; bacteriophage; capsule depoly-merase; capsule serotype; phage receptor, K2 serotype.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures
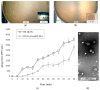


 ).
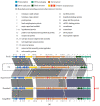
).



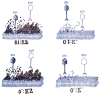
Similar articles
-
Isolation and Characterization of a Novel Lytic Bacteriophage against the K2 Capsule-Expressing Hypervirulent Klebsiella pneumoniae Strain 52145, and Identification of Its Functional Depolymerase.Microorganisms. 2021 Mar 21;9(3):650. doi: 10.3390/microorganisms9030650. Microorganisms. 2021. PMID: 33801047 Free PMC article.
-
A novel phage putative depolymerase, Depo16, has specific activity against K1 capsular-type Klebsiella pneumoniae.Appl Environ Microbiol. 2024 Apr 17;90(4):e0119723. doi: 10.1128/aem.01197-23. Epub 2024 Mar 29. Appl Environ Microbiol. 2024. PMID: 38551353 Free PMC article.
-
Comparison of the therapeutic potential of bacteriophage KpV74 and phage-derived depolymerase (β-glucosidase) against Klebsiella pneumoniae capsular type K2.Virus Res. 2022 Dec;322:198951. doi: 10.1016/j.virusres.2022.198951. Epub 2022 Sep 30. Virus Res. 2022. PMID: 36191686
-
Specificity and diversity of Klebsiella pneumoniae phage-encoded capsule depolymerases.Essays Biochem. 2024 Dec 17;68(5):661-677. doi: 10.1042/EBC20240015. Essays Biochem. 2024. PMID: 39668555 Free PMC article. Review.
-
Bacteriophage therapy: a possible alternative therapy against antibiotic-resistant strains of Klebsiella pneumoniae.Front Microbiol. 2025 Mar 31;16:1443430. doi: 10.3389/fmicb.2025.1443430. eCollection 2025. Front Microbiol. 2025. PMID: 40231234 Free PMC article. Review.
Cited by
-
Isolation and preliminary characterization of a novel bacteriophage vB_KquU_φKuK6 that infects the multidrug-resistant pathogen Klebsiella quasipneumoniae.Front Microbiol. 2024 Oct 15;15:1472729. doi: 10.3389/fmicb.2024.1472729. eCollection 2024. Front Microbiol. 2024. PMID: 39479209 Free PMC article.
-
A New Insight into Phage Combination Therapeutic Approaches Against Drug-Resistant Mixed Bacterial Infections.Phage (New Rochelle). 2024 Dec 18;5(4):203-222. doi: 10.1089/phage.2024.0011. eCollection 2024 Dec. Phage (New Rochelle). 2024. PMID: 40045937 Review.
-
Isolation and Characterization of a Novel Lytic Phage N22 and Its Effect on Drug-Resistant Klebsiella Pneumoniae.Infect Drug Resist. 2025 Apr 10;18:1807-1818. doi: 10.2147/IDR.S515363. eCollection 2025. Infect Drug Resist. 2025. PMID: 40231317 Free PMC article.
-
Genomic analysis and therapeutic efficacy evaluation of bacteriophage PK2420 for pneumonia caused by hypervirulent Klebsiella pneumoniae (K20 serotype).mSystems. 2025 May 20;10(5):e0163224. doi: 10.1128/msystems.01632-24. Epub 2025 Apr 16. mSystems. 2025. PMID: 40237452 Free PMC article.
-
Capsular Polysaccharide as a Potential Target in Hypervirulent and Drug-Resistant Klebsiella pneumoniae Treatment.Infect Drug Resist. 2025 Mar 3;18:1253-1262. doi: 10.2147/IDR.S493635. eCollection 2025. Infect Drug Resist. 2025. PMID: 40059940 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

