Genomic Analysis of the Deep-Sea Bacterium Shewanella sp. MTB7 Reveals Backgrounds Related to Its Deep-Sea Environment Adaptation
- PMID: 36985371
- PMCID: PMC10059138
- DOI: 10.3390/microorganisms11030798
Genomic Analysis of the Deep-Sea Bacterium Shewanella sp. MTB7 Reveals Backgrounds Related to Its Deep-Sea Environment Adaptation
Abstract
Shewanella species are widely distributed in various environments, especially deep-sea sediments, due to their remarkable ability to utilize multiple electron receptors and versatile metabolic capabilities. In this study, a novel facultatively anaerobic, psychrophilic, and piezotolerant bacterium, Shewanella sp. MTB7, was isolated from the Mariana Trench at a depth of 5900 m. Here, we report its complete genome sequence and adaptation strategies for survival in deep-sea environments. MTB7 contains what is currently the third-largest genome among all isolated Shewanella strains and shows higher coding density than neighboring strains. Metabolically, MTB7 is predicted to utilize various carbon and nitrogen sources. D-amino acid utilization and HGT-derived purine-degrading genes could contribute to its oligotrophic adaptation. For respiration, the cytochrome o ubiquinol oxidase genes cyoABCDE, typically expressed at high oxygen concentrations, are missing. Conversely, a series of anaerobic respiratory genes are employed, including fumarate reductase, polysulfide reductase, trimethylamine-N-oxide reductase, crotonobetaine reductase, and Mtr subunits. The glycine reductase genes and the triplication of dimethyl sulfoxide reductase genes absent in neighboring strains could also help MTB7 survive in low-oxygen environments. Many genes encoding cold-shock proteins, glycine betaine transporters and biosynthetic enzymes, and reactive oxygen species-scavenging proteins could contribute to its low-temperature adaptation. The genomic analysis of MTB7 will deepen our understanding of microbial adaptation strategies in deep-sea environments.
Keywords: Shewanella; complete genome; deep sea; psychrophilic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

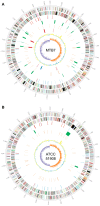
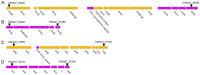
References
-
- Glud R.N., Berg P., Thamdrup B., Larsen M., Stewart H.A., Jamieson A.J., Glud A., Oguri K., Sanei H., Rowden A.A. Hadal trenches are dynamic hotspots for early diagenesis in the deep sea. Commun. Earth Environ. 2021;2:1–8. doi: 10.1038/s43247-020-00087-2. - DOI
-
- Inagaki F., Nunoura T., Nakagawa S., Teske A., Lever M., Lauer A., Suzuki M., Takai K., Delwiche M., Colwell F.S. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. USA. 2006;103:2815–2820. doi: 10.1073/pnas.0511033103. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

