Organic Anion Transporting Polypeptide 3A1 (OATP3A1)-Gated Bio-Orthogonal Labeling of Intracellular Proteins
- PMID: 36985493
- PMCID: PMC10055104
- DOI: 10.3390/molecules28062521
Organic Anion Transporting Polypeptide 3A1 (OATP3A1)-Gated Bio-Orthogonal Labeling of Intracellular Proteins
Abstract
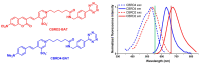
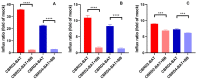
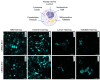
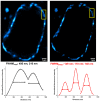
Organic anion transporting polypeptides (OATPs) were found to readily deliver membrane impermeable, tetrazine bearing fluorescent probes into cells. This feature was explored in OATP3A1 conditioned bio-orthogonal labeling schemes of various intracellular proteins in live cells. Confocal microscopy and super-resolution microscopy (STED) studies have shown that highly specific and efficient staining of the selected intracellular proteins can be achieved with the otherwise non-permeable probes when OATP3A1 is present in the cell membrane of cells. Such a transport protein linked bio-orthogonal labeling scheme is believed to be useful in OATP3A1 activity-controlled protein expression studies in the future.
Keywords: cell permeability; confocal microscopy; flow cytometry; intracellular labeling; large Stokes shift fluorescent probes; live cell bio-orthogonal modification; organic anion transporter polypeptide 3A1; self-labeling enzyme tags; super-resolution microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

