Novel Steroidal[17,16- d]pyrimidines Derived from Epiandrosterone and Androsterone: Synthesis, Characterization and Configuration-Activity Relationships
- PMID: 36985662
- PMCID: PMC10054084
- DOI: 10.3390/molecules28062691
Novel Steroidal[17,16- d]pyrimidines Derived from Epiandrosterone and Androsterone: Synthesis, Characterization and Configuration-Activity Relationships
Abstract
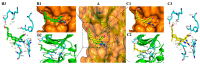
Two series of novel steroidal[17,16-d]pyrimidines derived from natural epiandrosterone and androsterone were designed and synthesized, and these compounds were screened for their potential anticancer activities. The preliminary bioassay indicated that some of these prepared compounds exhibited significantly good cytotoxic activities against human gastric cancer (SGC-7901), lung cancer (A549), and hepatocellular liver carcinoma (HepG2) cell lines compared with 5-fluorouracil (5-FU), epiandrosterone, and androsterone. Especially the respective pairs from epiandrosterone and androsterone showed significantly different inhibitory activities, and the possible configuration-activity relationships have also been summarized and discussed based on kinase assay and molecular docking, which indicated that the inhibition activities of these steroidal[17,16-d]pyrimidines might obviously be affected by the configuration of the hydroxyl group in the part of the steroidal scaffold.
Keywords: SARs; androsterone; bioactivity; epiandrosterone; molecular docking; steroidal[17,16-d]pyrimidines; synthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
-
- Mazumder K., Aktar A., Roy P., Biswas B., Hossain M., Sarkar K., Bachar S., Ahmed F., Monjur-Al-Hossain A., Fukase K. A Review on Mechanistic Insight of Plant Derived Anticancer Bioactive Phytocompounds and Their Structure Activity Relationship. Molecules. 2022;27:3036. doi: 10.3390/molecules27093036. - DOI - PMC - PubMed
-
- Song X.J., Shao Y., Dong X.G. Microwave-assisted synthesis of some novel fluorinated pyrazolo [3, 4-d] pyrimidine derivatives containing 1, 3, 4-thiadiazole as potential antitumor agents. Chin. Chem. Lett. 2011;22:1036–1038. doi: 10.1016/j.cclet.2011.05.012. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

