The Unusual Architecture of RNA-Dependent RNA Polymerase (RdRp)'s Catalytic Chamber Provides a Potential Strategy for Combination Therapy against COVID-19
- PMID: 36985777
- PMCID: PMC10057333
- DOI: 10.3390/molecules28062806
The Unusual Architecture of RNA-Dependent RNA Polymerase (RdRp)'s Catalytic Chamber Provides a Potential Strategy for Combination Therapy against COVID-19
Abstract
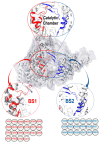
The unusual and interesting architecture of the catalytic chamber of the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) was recently explored using Cryogenic Electron Microscopy (Cryo-EM), which revealed the presence of two distinctive binding cavities within the catalytic chamber. In this report, first, we mapped out and fully characterized the variations between the two binding sites, BS1 and BS2, for significant differences in their amino acid architecture, size, volume, and hydrophobicity. This was followed by investigating the preferential binding of eight antiviral agents to each of the two binding sites, BS1 and BS2, to understand the fundamental factors that govern the preferential binding of each drug to each binding site. Results showed that, in general, hydrophobic drugs, such as remdesivir and sofosbuvir, bind better to both binding sites than relatively less hydrophobic drugs, such as alovudine, molnupiravir, zidovudine, favilavir, and ribavirin. However, suramin, which is a highly hydrophobic drug, unexpectedly showed overall weaker binding affinities in both binding sites when compared to other drugs. This unexpected observation may be attributed to its high binding solvation energy, which disfavors overall binding of suramin in both binding sites. On the other hand, hydrophobic drugs displayed higher binding affinities towards BS1 due to its higher hydrophobic architecture when compared to BS2, while less hydrophobic drugs did not show a significant difference in binding affinities in both binding sites. Analysis of binding energy contributions revealed that the most favorable components are the ΔEele, ΔEvdw, and ΔGgas, whereas ΔGsol was unfavorable. The ΔEele and ΔGgas for hydrophobic drugs were enough to balance the unfavorable ΔGsol, leaving the ΔEvdw to be the most determining factor of the total binding energy. The information presented in this report will provide guidelines for tailoring SARS-CoV-2 inhibitors with enhanced binding profiles.
Keywords: COVID-19; RNA-dependent RNA polymerase; catalytic chamber; combination therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Hasan A.H., Hussen N.H., Shakya S., Jamalis J., Pratama M.R.F., Chander S., Kharkwal H., Murugesan S. In silico discovery of multi-targeting inhibitors for the COVID-19 treatment by molecular docking, molecular dynamics simulation studies, and ADMET predictions. Struct. Chem. 2022;33:1645–1665. doi: 10.1007/s11224-022-01996-y. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

