Molecular Characterization of the First African Swine Fever Virus Genotype II Strains Identified from Mainland Italy, 2022
- PMID: 36986294
- PMCID: PMC10055901
- DOI: 10.3390/pathogens12030372
Molecular Characterization of the First African Swine Fever Virus Genotype II Strains Identified from Mainland Italy, 2022
Abstract
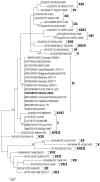
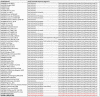
African swine fever (ASF) is responsible for important socio-economic effects in the global pig industry, especially for countries with large-scale piggery sectors. In January 2022, the African swine fever virus (ASFV) genotype II was identified in a wild boar population in mainland Italy (Piedmont region). This study describes the molecular characterization, by Sanger and next-generation sequencing (NGS), of the first index case 632/AL/2022 and of another isolate (2802/AL/2022) reported in the same month, in close proximity to the first, following multiple ASF outbreaks. Phylogenetic analysis based on the B646L gene and NGS clustered the isolates 632/AL/2022 and 2802/AL/2022 within the wide and most homogeneous p72 genotype II that includes viruses from European and Asian countries. The consensus sequence obtained from the ASFV 2802/AL/2022 isolate was 190,598 nucleotides in length and had a mean GC content of 38.38%. At the whole-genome level, ASF isolate 2802/AL/2022 showed a close genetic correlation with the other representative ASFV genotype II strains isolated between April 2007 and January 2022 from wild and domestic pigs in Eastern/Central European (EU) and Asian countries. CVR subtyping clustered the two Italian ASFV strains within the major CVR variant circulating since the first virus introduction in Georgia in 2007. Intergenic region I73R-I329L subtyping placed the Italian ASFV isolates within the variant identical to the strains frequently identified among wild boars and domestic pigs. Presently, given the high sequence similarity, it is impossible to trace the precise geographic origin of the virus at a country level. Moreover, the full-length sequences available in the NCBI are not completely representative of all affected territories.
Keywords: ASF outbreak; African swine fever virus (ASFV); genotype II; mainland Italy; next-generation sequencing (NGS); phylogenetic analysis; wild boar population.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be considered a potential conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript, nor in the decision to publish the results.
Figures


References
-
- Montgomery R.E. On a form of swine fever occurring in british east-Africa (Kenya Colony) J. Comp. Pathol. Ther. 1921;34:159–191. doi: 10.1016/S0368-1742(21)80031-4. - DOI
-
- Beltrán-Alcrudo D., Lubroth J., Depner K., De La Rocque S. African swine fever in the Caucasus. FAO Empres Watch. 2008;1:1–8. doi: 10.13140/RG.2.1.3579.1200. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

