A Review of Fatty Acid Biosynthesis Enzyme Inhibitors as Promising Antimicrobial Drugs
- PMID: 36986522
- PMCID: PMC10054515
- DOI: 10.3390/ph16030425
A Review of Fatty Acid Biosynthesis Enzyme Inhibitors as Promising Antimicrobial Drugs
Abstract
Resistance to antimicrobial drugs is currently a serious threat to human health. Consequently, we are facing an urgent need for new antimicrobial drugs acting with original modes of action. The ubiquitous and widely conserved microbial fatty acid biosynthesis pathway, called FAS-II system, represents a potential target to tackle antimicrobial resistance. This pathway has been extensively studied, and eleven proteins have been described. FabI (or InhA, its homologue in mycobacteria) was considered as a prime target by many teams and is currently the only enzyme with commercial inhibitor drugs: triclosan and isoniazid. Furthermore, afabicin and CG400549, two promising compounds which also target FabI, are in clinical assays to treat Staphylococcus aureus. However, most of the other enzymes are still underexploited targets. This review, after presenting the FAS-II system and its enzymes in Escherichia coli, highlights the reported inhibitors of the system. Their biological activities, main interactions formed with their targets and structure-activity relationships are presented as far as possible.
Keywords: FAS-II inhibitors; antimicrobial resistance; antimicrobials; fatty acid synthase system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








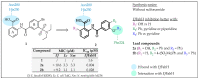
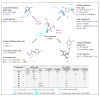





















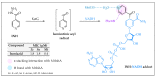







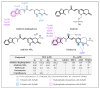




References
-
- World Health Organization The Top 10 Causes of Death. [(accessed on 27 February 2023)]. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death.
-
- Ikuta K.S., Swetschinski L.R., Aguilar G.R., Sharara F., Mestrovic T., Gray A.P., Weaver N.D., Wool E.E., Han C., Hayoon A.G., et al. Global Mortality Associated with 33 Bacterial Pathogens in 2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet. 2022;400:2221–2248. doi: 10.1016/S0140-6736(22)02185-7. - DOI - PMC - PubMed
-
- O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Wellcome Trust and UK Government; London, UK: 2016. Review on Antimicrobial Resistance.
-
- World Health Organization . World Malaria Report 2022. WHO; Geneva, Switzerland: 2022.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

