Current Status of Oligonucleotide-Based Protein Degraders
- PMID: 36986626
- PMCID: PMC10055846
- DOI: 10.3390/pharmaceutics15030765
Current Status of Oligonucleotide-Based Protein Degraders
Abstract
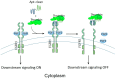
Transcription factors (TFs) and RNA-binding proteins (RBPs) have long been considered undruggable, mainly because they lack ligand-binding sites and are equipped with flat and narrow protein surfaces. Protein-specific oligonucleotides have been harnessed to target these proteins with some satisfactory preclinical results. The emerging proteolysis-targeting chimera (PROTAC) technology is no exception, utilizing protein-specific oligonucleotides as warheads to target TFs and RBPs. In addition, proteolysis by proteases is another type of protein degradation. In this review article, we discuss the current status of oligonucleotide-based protein degraders that are dependent either on the ubiquitin-proteasome system or a protease, providing a reference for the future development of degraders.
Keywords: PROTAC; SNIPER; chimera; degrader therapeutics; nucleic acids; proteases; targeted protein degradation.
Conflict of interest statement
M.N. (Mikihiko Naito) belongs to the Social Cooperation Program of Targeted Protein Degradation supported by Eisai Co., Ltd. and serves as a scientific advisor to UBiENCE Inc. The other authors declare no conflict of interest.
Figures



References
Publication types
Grants and funding
- JP18H05502/Japan Society for the Promotion of Science
- JP21H02777/Japan Society for the Promotion of Science
- JP22F22107/Japan Society for the Promotion of Science
- JP21K05320/Japan Society for the Promotion of Science
- P22107/Japan Society for the Promotion of Science
- 2121J23036/Japan Society for the Promotion of Science
- JP22ym0126805/Japan Agency for Medical Research and Development
- JP21mk0101197/Japan Agency for Medical Research and Development
- JP21ak0101073/Japan Agency for Medical Research and Development
- XXX/Princess Takamatsu Cancer Research Fund
- XXX/Terumo Foundation for Life Sciences and Arts
- XXX/Takeda Science Foundation
- XXX/Naito Foundation
- XXX/Sumitomo Foundation
- XXX/the Novartis Foundation (Japan) for the Promotion of Science
- XXX/Japan Foundation of Applied Enzymology
- XXX/Kobayashi Foundation for Cancer Research
- XXX/Foundation for Promotion of Cancer Research in Japan
- XXX/Tokyo Biochemical Research Foundation
LinkOut - more resources
Full Text Sources
Other Literature Sources

