Identification and Characterization of Common Bean (Phaseolus vulgaris) Non-Nodulating Mutants Altered in Rhizobial Infection
- PMID: 36986997
- PMCID: PMC10059843
- DOI: 10.3390/plants12061310
Identification and Characterization of Common Bean (Phaseolus vulgaris) Non-Nodulating Mutants Altered in Rhizobial Infection
Abstract
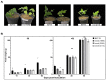
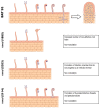
The symbiotic N2-fixation process in the legume-rhizobia interaction is relevant for sustainable agriculture. The characterization of symbiotic mutants, mainly in model legumes, has been instrumental for the discovery of symbiotic genes, but similar studies in crop legumes are scant. To isolate and characterize common bean (Phaseolus vulgaris) symbiotic mutants, an ethyl methanesulphonate-induced mutant population from the BAT 93 genotype was analyzed. Our initial screening of Rhizobium etli CE3-inoculated mutant plants revealed different alterations in nodulation. We proceeded with the characterization of three non-nodulating (nnod), apparently monogenic/recessive mutants: nnod(1895), nnod(2353) and nnod(2114). Their reduced growth in a symbiotic condition was restored when the nitrate was added. A similar nnod phenotype was observed upon inoculation with other efficient rhizobia species. A microscopic analysis revealed a different impairment for each mutant in an early symbiotic step. nnod(1895) formed decreased root hair curling but had increased non-effective root hair deformation and no rhizobia infection. nnod(2353) produced normal root hair curling and rhizobia entrapment to form infection chambers, but the development of the latter was blocked. nnod(2114) formed infection threads that did not elongate and thus did not reach the root cortex level; it occasionally formed non-infected pseudo-nodules. The current research is aimed at mapping the responsible mutated gene for a better understanding of SNF in this critical food crop.
Keywords: EMS; Phaseolus vulgaris; infection; nodulation; rhizobia; symbiosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Broughton W.J., Hernández G., Blair M., Beebe S., Gepts P., Vanderleyden J. Beans (Phaseolus Spp.)–Model Food Legumes. Plant Soil. 2003;252:55–128. doi: 10.1023/A:1024146710611. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

