Evolutionary Analysis of the Melon (Cucumis melo L.) GH3 Gene Family and Identification of GH3 Genes Related to Fruit Growth and Development
- PMID: 36987071
- PMCID: PMC10053650
- DOI: 10.3390/plants12061382
Evolutionary Analysis of the Melon (Cucumis melo L.) GH3 Gene Family and Identification of GH3 Genes Related to Fruit Growth and Development
Abstract
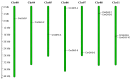
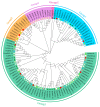
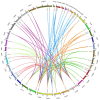
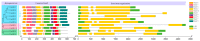
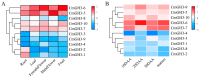
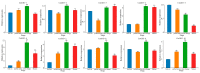
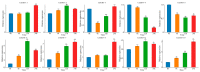
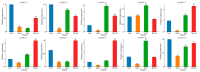
The indole-3-acetic acid (IAA) auxin is an important endogenous hormone that plays a key role in the regulation of plant growth and development. In recent years, with the progression of auxin-related research, the function of the Gretchen Hagen 3 (GH3) gene has become a prominent research topic. However, studies focusing on the characteristics and functions of melon GH3 family genes are still lacking. This study presents a systematic identification of melon GH3 gene family members based on genomic data. The evolution of melon GH3 family genes was systematically analyzed by means of bioinformatics, and the expression patterns of the GH3 family genes in different melon tissues during different fruit developmental stages and with various levels of 1-naphthaleneacetic acid (NAA) induction were analyzed with transcriptomics and RT-qPCR. The melon genome contains 10 GH3 genes distributed across seven chromosomes, and most of these genes are expressed in the plasma membrane. According to evolutionary analysis and the number of GH3 family genes, these genes can be divided into three subgroups, and they have been conserved throughout the evolution of melon. The melon GH3 gene has a wide range of expression patterns across distinct tissue types, with expression generally being higher in flowers and fruit. Through promoter analysis, we found that most cis-acting elements contained light- and IAA-responsive elements. Based on the RNA-seq and RT-qPCR analyses, it can be speculated that CmGH3-5, CmGH3-6 and CmGH3-7 may be involved in the process of melon fruit development. In conclusion, our findings suggest that the GH3 gene family plays an important role in the development of melon fruit. This study provides an important theoretical basis for further research on the function of the GH3 gene family and the molecular mechanism underlying the development of melon fruit.
Keywords: GH3 gene family; evolutionary analysis; expression analysis; fruit development; melon.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures

Similar articles
-
Genome-wide identification and expression analysis of the JMJ-C gene family in melon (Cucumis melo L.) reveals their potential role in fruit development.BMC Genomics. 2023 Dec 13;24(1):771. doi: 10.1186/s12864-023-09868-3. BMC Genomics. 2023. PMID: 38093236 Free PMC article.
-
Genome-wide characterization and expression analysis of SAUR gene family in Melon (Cucumis melo L.).Planta. 2022 May 13;255(6):123. doi: 10.1007/s00425-022-03908-0. Planta. 2022. PMID: 35552537
-
Genome-wide analysis of cotton GH3 subfamily II reveals functional divergence in fiber development, hormone response and plant architecture.BMC Plant Biol. 2018 Dec 12;18(1):350. doi: 10.1186/s12870-018-1545-5. BMC Plant Biol. 2018. PMID: 30541440 Free PMC article.
-
The Roles of GRETCHEN HAGEN3 (GH3)-Dependent Auxin Conjugation in the Regulation of Plant Development and Stress Adaptation.Plants (Basel). 2023 Dec 8;12(24):4111. doi: 10.3390/plants12244111. Plants (Basel). 2023. PMID: 38140438 Free PMC article. Review.
-
Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function.Int J Mol Sci. 2018 Jan 16;19(1):259. doi: 10.3390/ijms19010259. Int J Mol Sci. 2018. PMID: 29337875 Free PMC article. Review.
Cited by
-
Genome-wide identification and expression characterization of the GH3 gene family of tea plant (Camellia sinensis).BMC Genomics. 2024 Jan 27;25(1):120. doi: 10.1186/s12864-024-10004-y. BMC Genomics. 2024. PMID: 38280985 Free PMC article.
-
Transcriptomic and Metabolomic Joint Analysis Revealing Different Metabolic Pathways and Genes Dynamically Regulating Bitter Gourd (Momordica charantia L.) Fruit Growth and Development in Different Stages.Plants (Basel). 2025 Jul 21;14(14):2248. doi: 10.3390/plants14142248. Plants (Basel). 2025. PMID: 40733485 Free PMC article.
-
Comprehensive analysis of transcriptome and metabolome identified the key gene networks regulating fruit length in melon.BMC Plant Biol. 2025 Apr 8;25(1):442. doi: 10.1186/s12870-025-06332-0. BMC Plant Biol. 2025. PMID: 40200143 Free PMC article.
-
GH3 Gene Family Identification in Chinese White Pear (Pyrus bretschneideri) and the Functional Analysis of PbrGH3.5 in Fe Deficiency Responses in Tomato.Int J Mol Sci. 2024 Dec 3;25(23):12980. doi: 10.3390/ijms252312980. Int J Mol Sci. 2024. PMID: 39684691 Free PMC article.
-
A Scoping Review on Cucumis melo and Its Anti-Cancer Properties.Malays J Med Sci. 2024 Aug;31(4):63-77. doi: 10.21315/mjms2024.31.4.5. Epub 2024 Aug 27. Malays J Med Sci. 2024. PMID: 39247112 Free PMC article.
References
-
- Flores-León A., García-Martínez S., González V., Garcés-Claver A., Martí R., Julián C., Sifres A., Pérez-de-Castro A., Díez M.J., López C., et al. Grafting Snake Melon [Cucumis melo L. subsp. Melo Var. Flexuosus (L.) Naudin] in Organic Farming: Effects on Agronomic Performance; Resistance to Pathogens; Sugar, Acid, and VOC Profiles; and Consumer Acceptance. Front. Plant Sci. 2021;12:613845. doi: 10.3389/fpls.2021.613845. - DOI - PMC - PubMed
-
- Zhang Z., Zhang Z., Han X., Wu J., Zhang L., Wang J., Wang-Pruski G. Specific response mechanism to autotoxicity in melon (Cucumis melo L.) root revealed by physiological analyses combined with transcriptome profiling. Ecotoxicol. Environ. Saf. 2020;200:110779. doi: 10.1016/j.ecoenv.2020.110779. - DOI - PubMed
-
- Lopes A.P., Galuch M.B., Petenuci M.E., Oliveira J.H., Canesin E.A., Schneider V.V.A., Visentainer J.V. Quantification of phenolic compounds in ripe and unripe bitter melons (Momordica charantia) and evaluation of the distribution of phenolic compounds in different parts of the fruit by UPLC–MS/MS. Chem. Pap. 2020;74:2613–2625. doi: 10.1007/s11696-020-01094-5. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

