H4C5 missense variant leads to a neurodevelopmental phenotype overlapping with Angelman syndrome
- PMID: 36987712
- PMCID: PMC10286100
- DOI: 10.1002/ajmg.a.63193
H4C5 missense variant leads to a neurodevelopmental phenotype overlapping with Angelman syndrome
Abstract
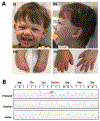
Recurrent de novo missense variants in H4 histone genes have recently been associated with a novel neurodevelopmental syndrome that is characterized by intellectual disability and developmental delay as well as more variable findings that include short stature, microcephaly, and facial dysmorphisms. A 4-year-old male with autism, developmental delay, microcephaly, and a happy demeanor underwent evaluation through the Undiagnosed Disease Network. He was clinically suspected to have Angelman syndrome; however, molecular testing was negative. Genome sequencing identified the H4 histone gene variant H4C5 NM_003545.4: c.295T>C, p.Tyr99His, which parental testing confirmed to be de novo. The variant met criteria for a likely pathogenic classification and is one of the seven known disease-causing missense variants in H4C5. A comparison of our proband's findings to the initial description of the H4-associated neurodevelopmental syndrome demonstrates that his phenotype closely matches the spectrum of those reported among the 29 affected individuals. As such, this report corroborates the delineation of neurodevelopmental syndrome caused by de novo missense H4 gene variants. Moreover, it suggests that cases of clinically suspected Angelman syndrome without molecular confirmation should undergo exome or genome sequencing, as novel neurodevelopmental syndromes with phenotypes overlapping with Angelman continue to be discovered.
Keywords: Angelman syndrome; H4C5; digit anomalies; microcephaly; neurodevelopmental syndrome; tooth anomalies.
© 2023 Wiley Periodicals LLC.
Conflict of interest statement
Conflict of Interest
The authors have no conflicts of interest to report.
Figures
References
-
- Bryant L, Li D, Cox SG, Marchione D, Joiner EF, Wilson K, Janssen K, Lee P, March ME, Nair D, Sherr E, Fregeau B, Wierenga KJ, Wadley A, Mancini GMS, Powell-Hamilton N, Kamp J. van de, Grebe T, Dean J, … Bhoj EJ (2020). Histone H3.3 beyond cancer: Germline mutations in Histone 3 Family 3A and 3B cause a previously unidentified neurodegenerative disorder in 46 patients. Science Advances, 6(49), eabc9207. 10.1126/sciadv.abc9207 - DOI - PMC - PubMed
-
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Angel G. del, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, & Daly MJ. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics, 43(5), 491–498. 10.1038/ng.806 - DOI - PMC - PubMed
-
- Ewans LJ, Schofield D, Shrestha R, Zhu Y, Gayevskiy V, Ying K, Walsh C, Lee E, Kirk EP, Colley A, Ellaway C, Turner A, Mowat D, Worgan L, Freckmann M-L, Lipke M, Sachdev R, Miller D, Field M, … Roscioli T. (2018). Whole-exome sequencing reanalysis at 12 months boosts diagnosis and is cost-effective when applied early in Mendelian disorders. Genetics in Medicine, 20(12), 1564–1574. 10.1038/gim.2018.39 - DOI - PubMed
-
- Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A, Li Q, Holzinger E, Karyadi D, Cannon-Albright LA, Teerlink CC, Stanford JL, Isaacs WB, Xu J, Cooney KA, Lange EM, Schleutker J, Carpten JD, … Sieh W (2016). REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. The American Journal of Human Genetics, 99(4), 877–885. 10.1016/j.ajhg.2016.08.016 - DOI - PMC - PubMed
-
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, … MacArthur DG (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature, 581(7809), 434–443. 10.1038/s41586-020-2308-7 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

