Phenotypic and transcriptomic impact of expressing mammalian TET2 in the Drosophila melanogaster model
- PMID: 36989121
- PMCID: PMC10072067
- DOI: 10.1080/15592294.2023.2192375
Phenotypic and transcriptomic impact of expressing mammalian TET2 in the Drosophila melanogaster model
Abstract
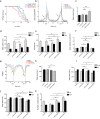
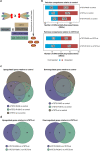
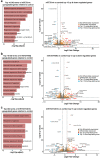
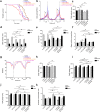
Ten-Eleven Translocation (TET) proteins have recently come to light as important epigenetic regulators conserved in multicellular organisms. TET knockdown studies in rodents have highlighted the critical role of these proteins for proper brain development and function. Mutations in mammalian mTET proteins and mTET2 specifically are frequent and deregulated in leukaemia and glioma respectively. Accordingly, we examined the role of mTET2 in tumorigenesis in larval haemocytes and adult heads in Drosophila melanogaster. Our findings showed that expression of mutant and wild type mTET2 resulted in general phenotypic defects in adult flies and accumulation of abdominal melanotic masses. Notably, flies with mTET2-R43G mutation at the N-terminus of mTET2 exhibited locomotor and circadian behavioural deficits, as well as reduced lifespan. Flies with mTET2-R1261C mutation in the catalytic domain, a common mutation in acute myeloid leukaemia (AML), displayed alterations affecting haemocyte haemostasis. Using transcriptomic approach, we identified upregulated immune genes in fly heads that were not exclusive to TET2 mutants but also found in wild type mTET2 flies. Furthermore, inhibiting expression of genes that were found to be deregulated in mTET2 mutants, such as those involved in immune pathways, autophagy, and transcriptional regulation, led to a rescue in fly survival, behaviour, and hemocyte number. This study identifies the transcriptomic profile of wild type mTET2 versus mTET2 mutants (catalytic versus non-catalytic) with indications of TET2 role in normal central nervous system (CNS) function and innate immunity.
Keywords: Drosophila melanogaster; TET2; behaviour; circadian rhythm; glioma; innate immunity; leukaemia.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
