Cholinergic regulation of vascular endothelial function by human ChAT+ T cells
- PMID: 36989306
- PMCID: PMC10083572
- DOI: 10.1073/pnas.2212476120
Cholinergic regulation of vascular endothelial function by human ChAT+ T cells
Abstract
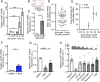
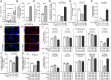
Endothelial dysfunction and impaired vasodilation are linked with adverse cardiovascular events. T lymphocytes expressing choline acetyltransferase (ChAT), the enzyme catalyzing biosynthesis of the vasorelaxant acetylcholine (ACh), regulate vasodilation and are integral to the cholinergic antiinflammatory pathway in an inflammatory reflex in mice. Here, we found that human T cell ChAT mRNA expression was induced by T cell activation involving the PI3K signaling cascade. Mechanistically, we identified that ChAT mRNA expression was induced following the attenuation of RE-1 Silencing Transcription factor REST-mediated methylation of the ChAT promoter, and that ChAT mRNA expression levels were up-regulated by GATA3 in human T cells. In functional experiments, T cell-derived ACh increased endothelial nitric oxide-synthase activity, promoted vasorelaxation, and reduced vascular endothelial activation and promoted barrier integrity by a cholinergic mechanism. Further, we observed that survival in a cohort of patients with severe circulatory failure correlated with their relative frequency of ChAT +CD4+ T cells in blood. These findings on ChAT+ human T cells provide a mechanism for cholinergic immune regulation of vascular endothelial function in human inflammation.
Keywords: acetylcholine; circulation; lymphocytes; vascular biology.
Conflict of interest statement
P.S.O. is a co-owner of Emune AB., P.S.O. is listed as a co-inventor on a patent regarding potential therapeutic use of ChAT+ T cells. M.E. has received honoraria for lectures and consultancy from AbbVie, Merck Sharp and Dohme, Takeda, Ferring, Orion Pharma, Otsuka, Tillotts, Immune Therapy Holdings, Novartis, Pfizer, Bristol Myers Squibb, and Janssen and received research funding from AbbVie and Merck Sharp and Dohme.
Figures



Comment in
-
Acetylcholine-producing CD4 T cells regulate vasculature in humans.Proc Natl Acad Sci U S A. 2023 Apr 18;120(16):e2303525120. doi: 10.1073/pnas.2303525120. Epub 2023 Apr 10. Proc Natl Acad Sci U S A. 2023. PMID: 37036991 Free PMC article. No abstract available.
References
-
- Cines D. B., et al. , Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood 91, 3527–3561 (1998). - PubMed
-
- Bonetti P. O., Lerman L. O., Lerman A., Endothelial dysfunction. Arterioscler. Thromb. Vasc. Biol. 23, 168–175 (2003). - PubMed
-
- Vincent J.-L., De Backer D., Circulatory shock. N. Engl. J. Med. 369, 1726–1734 (2013). - PubMed
-
- Harjola V. P., et al. , Clinical picture and risk prediction of short-term mortality in cardiogenic shock. Eur. J. Heart Fail. 17, 501–509 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

