Tubastatin A potently inhibits GPX4 activity to potentiate cancer radiotherapy through boosting ferroptosis
- PMID: 36989572
- PMCID: PMC10074938
- DOI: 10.1016/j.redox.2023.102677
Tubastatin A potently inhibits GPX4 activity to potentiate cancer radiotherapy through boosting ferroptosis
Abstract
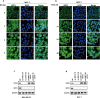
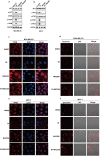
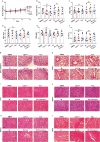
Ferroptosis, an iron-dependent lipid peroxidation-driven programmed cell death, is closely related to cancer therapy. The development of druggable ferroptosis inducers and their rational application in cancer therapy are critical. Here, we identified Tubastatin A, an HDAC6 inhibitor as a novel druggable ferroptosis inducer through large-scale drug screening. Tubastatin A directly bonded to GPX4 and inhibited GPX4 enzymatic activity through biotin-linked Tubastatin A putdown and LC/MS analysis, which is independent of its inhibition of HDAC6. In addition, our results showed that radiotherapy not only activated Nrf2-mediated GPX4 transcription but also inhibited lysosome-mediated GPX4 degradation, subsequently inducing ferroptosis tolerance and radioresistance in cancer cells. Tubastatin A overcame ferroptosis resistance and radioresistance of cancer cells by inhibiting GPX4 enzymatic activity. More importantly, Tubastatin A has excellent bioavailability, as demonstrated by its ability to significantly promote radiotherapy-induced lipid peroxidation and tumour suppression in a mouse xenograft model. Our findings identify a novel druggable ferroptosis inducer, Tubastatin A, which enhances radiotherapy-mediated antitumor effects. This work provides a compelling rationale for the clinical evaluation of Tubastatin A, especially in combination with radiotherapy.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


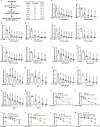
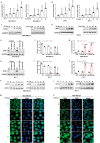


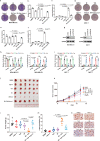
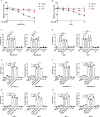


References
-
- Hassannia B., Vandenabeele P., Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35:830–849. - PubMed
-
- Zhang H.L., Hu B.X., Li Z.L., Du T., Shan J.L., Ye Z.P., Peng X.D., Li X., Huang Y., Zhu X.Y., Chen Y.H., Feng G.K., Yang D., Deng R., Zhu X.F. PKCbetaII phosphorylates ACSL4 to amplify lipid peroxidation to induce ferroptosis. Nat. Cell Biol. 2022;24:88–98. - PubMed
-
- Yan B., Ai Y., Sun Q., Ma Y., Cao Y., Wang J., Zhang Z., Wang X. Membrane damage during ferroptosis is caused by oxidation of phospholipids catalyzed by the oxidoreductases POR and CYB5R1. Mol. Cell. 2021;81:355–369 e310. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

