Pathologically high intraocular pressure induces mitochondrial dysfunction through Drp1 and leads to retinal ganglion cell PANoptosis in glaucoma
- PMID: 36989574
- PMCID: PMC10074988
- DOI: 10.1016/j.redox.2023.102687
Pathologically high intraocular pressure induces mitochondrial dysfunction through Drp1 and leads to retinal ganglion cell PANoptosis in glaucoma
Abstract
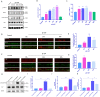
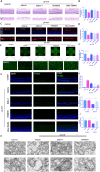
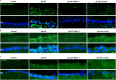
Glaucoma is a common neurodegenerative disease characterized by progressive retinal ganglion cell (RGC) loss and visual field defects. Pathologically high intraocular pressure (ph-IOP) is an important risk factor for glaucoma, and it triggers molecularly distinct cascades that control RGC death and axonal degeneration. Dynamin-related protein 1 (Drp1)-mediated abnormalities in mitochondrial dynamics are involved in glaucoma pathogenesis; however, little is known about the precise pathways that regulate RGC injury and death. Here, we aimed to investigate the role of the ERK1/2-Drp1-reactive oxygen species (ROS) axis in RGC death and the relationship between Drp1-mediated mitochondrial dynamics and PANoptosis in ph-IOP injury. Our results suggest that inhibiting the ERK1/2-Drp1-ROS pathway is a potential therapeutic strategy for treating ph-IOP-induced injuries. Furthermore, inhibiting Drp1 can regulate RGC PANoptosis by modulating caspase3-dependent, nucleotide-binding oligomerization domain-like receptor-containing pyrin domain 3(NLRP3)-dependent, and receptor-interacting protein (RIP)-dependent pathways in the ph-IOP model. Overall, our findings provide new insights into possible protective interventions that could regulate mitochondrial dynamics to improve RGC survival.
Keywords: Dynamin-related protein 1; Glaucoma; Mitochondrial dynamics; PANoptosis; Retinal ganglion cell.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
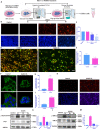
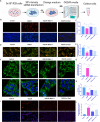
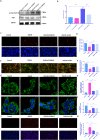
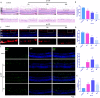


References
-
- Kang J.M., Tanna A.P. Glaucoma. Med. Clin. 2021;105:493–510. - PubMed
-
- Almasieh M., Wilson A.M., Morquette B., Cueva Vargas J.L., Di Polo A. The molecular basis of retinal ganglion cell death in glaucoma. Prog. Retin. Eye Res. 2012;31:152–181. - PubMed
-
- Sun M.H., Pang J.H., Chen S.L., Han W.H., Ho T.C., Chen K.J., Kao L.Y., Lin K.K., Tsao Y.P. Retinal protection from acute glaucoma-induced ischemia-reperfusion injury through pharmacologic induction of heme oxygenase-1. Invest. Ophthalmol. Vis. Sci. 2010;51:4798–4808. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

