Genome-wide association study of obstructive sleep apnoea in the Million Veteran Program uncovers genetic heterogeneity by sex
- PMID: 36989840
- PMCID: PMC10065974
- DOI: 10.1016/j.ebiom.2023.104536
Genome-wide association study of obstructive sleep apnoea in the Million Veteran Program uncovers genetic heterogeneity by sex
Abstract
Background: Genome-wide association studies (GWAS) for obstructive sleep apnoea (OSA) are limited due to the underdiagnosis of OSA, leading to misclassification of OSA, which consequently reduces statistical power. We performed a GWAS of OSA in the Million Veteran Program (MVP) of the U.S. Department of Veterans Affairs (VA) healthcare system, where OSA prevalence is close to its true population prevalence.
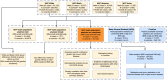
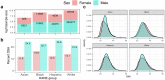
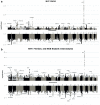
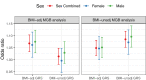
Methods: We performed GWAS of 568,576 MVP participants, stratified by biological sex and by harmonized race/ethnicity and genetic ancestry (HARE) groups of White, Black, Hispanic, and Asian individuals. We considered both BMI adjusted (BMI-adj) and unadjusted (BMI-unadj) models. We replicated associations in independent datasets, and analysed the heterogeneity of OSA genetic associations across HARE and sex groups. We finally performed a larger meta-analysis GWAS of MVP, FinnGen, and the MGB Biobank, totalling 916,696 individuals.
Findings: MVP participants are 91% male. OSA prevalence is 21%. In MVP there were 18 and 6 genome-wide significant loci in BMI-unadj and BMI-adj analyses, respectively, corresponding to 21 association regions. Of these, 17 were not previously reported in association with OSA, and 13 replicated in FinnGen (False Discovery Rate p-value < 0.05). There were widespread significant differences in genetic effects between men and women, but less so across HARE groups. Meta-analysis of MVP, FinnGen, and MGB biobank revealed 17 additional, previously unreported, genome-wide significant regions.
Interpretation: Sex differences in genetic associations with OSA are widespread, likely associated with multiple OSA risk factors. OSA shares genetic underpinnings with several sleep phenotypes, suggesting shared aetiology and causal pathways.
Funding: Described in acknowledgements.
Keywords: Electronic health records; Genome-wide association study; Obstructive sleep apnoea; Sex differences.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests DJ Gottlieb reports receiving support from NIH grants R01HL137234, R01 NR018335, and R01HL153874, and VA/DOE grant MVP063 (service ID #), 826-PS-null-45187. Also, he reports participation in a data safety monitoring board or an advisory board for Signifier Medical Technologies, Inc, Wesper, Inc, and Powell–Mansfield, Inc. E Abner reports receiving support from Estonian Research Council grant nr. MOBERA21, and NIH grant nr. 2R01DK075787-11. T Sofer reports receiving support from the National Heart Lung and Blood Institute, with payments made to the institution. T Esko reports receiving support from Estonian Research Council grant nr. MOBERA21, and NIH grant nr. 2R01DK075787-11.
Figures


References
-
- Chan A.S.L., Phillips C.L., Cistulli P.A. Obstructive sleep apnoea--an update. Intern Med J. 2010;40(2):102–106. - PubMed
-
- de Paula L.K.G., Alvim R.O., Pedrosa R.P., et al. Heritability of OSA in a rural population. Chest. 2016;149(1):92–97. - PubMed
-
- Redline S., Tishler P.V., Tosteson T.D., et al. The familial aggregation of obstructive sleep apnea. Am J Respir Crit Care Med. 1995;151(3 Pt 1):682–687. - PubMed
-
- Redline S., Tishler P.V. The genetics of sleep apnea. Sleep Med Rev. 2000;4(6):583–602. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

