Spatial multiomics map of trophoblast development in early pregnancy
- PMID: 36991123
- PMCID: PMC10076224
- DOI: 10.1038/s41586-023-05869-0
Spatial multiomics map of trophoblast development in early pregnancy
Abstract
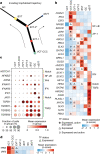
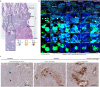
The relationship between the human placenta-the extraembryonic organ made by the fetus, and the decidua-the mucosal layer of the uterus, is essential to nurture and protect the fetus during pregnancy. Extravillous trophoblast cells (EVTs) derived from placental villi infiltrate the decidua, transforming the maternal arteries into high-conductance vessels1. Defects in trophoblast invasion and arterial transformation established during early pregnancy underlie common pregnancy disorders such as pre-eclampsia2. Here we have generated a spatially resolved multiomics single-cell atlas of the entire human maternal-fetal interface including the myometrium, which enables us to resolve the full trajectory of trophoblast differentiation. We have used this cellular map to infer the possible transcription factors mediating EVT invasion and show that they are preserved in in vitro models of EVT differentiation from primary trophoblast organoids3,4 and trophoblast stem cells5. We define the transcriptomes of the final cell states of trophoblast invasion: placental bed giant cells (fused multinucleated EVTs) and endovascular EVTs (which form plugs inside the maternal arteries). We predict the cell-cell communication events contributing to trophoblast invasion and placental bed giant cell formation, and model the dual role of interstitial EVTs and endovascular EVTs in mediating arterial transformation during early pregnancy. Together, our data provide a comprehensive analysis of postimplantation trophoblast differentiation that can be used to inform the design of experimental models of the human placenta in early pregnancy.
© 2023. The Author(s).
Conflict of interest statement
S.A.T. has received remuneration for consulting and scientific advisory board work from Genentech, Biogen, Roche and GlaxoSmithKline as well as Foresite Labs over the past three years. O.S. is a paid scientific advisory board member of Insitro Inc.
Figures















Comment in
-
Understanding human placentation through spatial multiomics.Nat Rev Genet. 2023 Jun;24(6):347. doi: 10.1038/s41576-023-00607-8. Nat Rev Genet. 2023. PMID: 37076575 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

