STING inhibits the reactivation of dormant metastasis in lung adenocarcinoma
- PMID: 36991128
- PMCID: PMC10569211
- DOI: 10.1038/s41586-023-05880-5
STING inhibits the reactivation of dormant metastasis in lung adenocarcinoma
Abstract
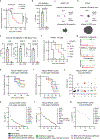
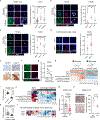
Metastasis frequently develops from disseminated cancer cells that remain dormant after the apparently successful treatment of a primary tumour. These cells fluctuate between an immune-evasive quiescent state and a proliferative state liable to immune-mediated elimination1-6. Little is known about the clearing of reawakened metastatic cells and how this process could be therapeutically activated to eliminate residual disease in patients. Here we use models of indolent lung adenocarcinoma metastasis to identify cancer cell-intrinsic determinants of immune reactivity during exit from dormancy. Genetic screens of tumour-intrinsic immune regulators identified the stimulator of interferon genes (STING) pathway as a suppressor of metastatic outbreak. STING activity increases in metastatic progenitors that re-enter the cell cycle and is dampened by hypermethylation of the STING promoter and enhancer in breakthrough metastases or by chromatin repression in cells re-entering dormancy in response to TGFβ. STING expression in cancer cells derived from spontaneous metastases suppresses their outgrowth. Systemic treatment of mice with STING agonists eliminates dormant metastasis and prevents spontaneous outbreaks in a T cell- and natural killer cell-dependent manner-these effects require cancer cell STING function. Thus, STING provides a checkpoint against the progression of dormant metastasis and a therapeutically actionable strategy for the prevention of disease relapse.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
SWL received funding and research support from Agilent Technologies for the purposes of massively parallel oligo synthesis to generate the sgRNA libraries described herein.
Figures


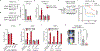









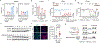
References
-
- Pantel K et al. Frequent down-regulation of major histocompatibility class I antigen expression on individual micrometastatic carcinoma cells. Cancer Res 51, 4712–4715 (1991). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

