Genetically engineered CXCR4-modified exosomes for delivery of miR-126 mimics to macrophages alleviate periodontitis
- PMID: 36991451
- PMCID: PMC10061745
- DOI: 10.1186/s12951-023-01863-w
Genetically engineered CXCR4-modified exosomes for delivery of miR-126 mimics to macrophages alleviate periodontitis
Abstract
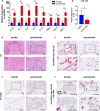
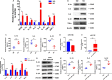
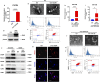
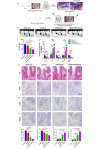
Biofilm-related diseases are a group of diseases that tolerate antimicrobial chemotherapies and therefore are refractory to treatment. Periodontitis, a non-device chronic biofilm disease induced by dental plaque, can serve as an excellent in vivo model to study the important effects of host factors on the biofilm microenvironment. Macrophage activity is one of the key factors that modulate the progression of inflammation-driven destruction in periodontitis; therefore it is an important host immunomodulatory factor. In this study, the reduction of microRNA-126 (miR-126) with the recruitment of macrophages in periodontitis was confirmed in clinical samples, and a strategy for targeted delivery of miR-126 to macrophages was explored. Exosomes overexpressing the C-X-C motif chemokine receptor 4 (CXCR4) loaded with miR-126 (CXCR4-miR126-Exo) was successfully constructed, which reduced off-target delivery to macrophages and regulated macrophages toward the anti-inflammatory phenotype. In vivo local injection of CXCR4-miR126-Exo into sites of periodontitis in rats effectively reduced bone resorption and osteoclastogenesis and inhibited the progression of periodontitis. These results provide new insights for designing novel immunomodulatory factor targeted delivery systems to treat periodontitis and other biofilm-related diseases.
Keywords: Biofilm diseases; Extracellular vesicles; Gene therapy; Immunomodulation; Targeted delivery.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Akcalı A, Lang NP. Dental calculus: the calcified biofilm and its role in disease development. Periodontol 2000. 2018;76:109–15. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

