Combined Use of RT-qPCR and NGS for Identification and Surveillance of SARS-CoV-2 Variants of Concern in Residual Clinical Laboratory Samples in Miami-Dade County, Florida
- PMID: 36992302
- PMCID: PMC10059866
- DOI: 10.3390/v15030593
Combined Use of RT-qPCR and NGS for Identification and Surveillance of SARS-CoV-2 Variants of Concern in Residual Clinical Laboratory Samples in Miami-Dade County, Florida
Abstract
Over the course of the COVID-19 pandemic, SARS-CoV-2 variants of concern (VOCs) with increased transmissibility and immune escape capabilities, such as Delta and Omicron, have triggered waves of new COVID-19 infections worldwide, and Omicron subvariants continue to represent a global health concern. Tracking the prevalence and dynamics of VOCs has clinical and epidemiological significance and is essential for modeling the progression and evolution of the COVID-19 pandemic. Next generation sequencing (NGS) is recognized as the gold standard for genomic characterization of SARS-CoV-2 variants, but it is labor and cost intensive and not amenable to rapid lineage identification. Here we describe a two-pronged approach for rapid, cost-effective surveillance of SARS-CoV-2 VOCs by combining reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) and periodic NGS with the ARTIC sequencing method. Variant surveillance by RT-qPCR included the commercially available TaqPath COVID-19 Combo Kit to track S-gene target failure (SGTF) associated with the spike protein deletion H69-V70, as well as two internally designed and validated RT-qPCR assays targeting two N-terminal-domain (NTD) spike gene deletions, NTD156-7 and NTD25-7. The NTD156-7 RT-qPCR assay facilitated tracking of the Delta variant, while the NTD25-7 RT-qPCR assay was used for tracking Omicron variants, including the BA.2, BA.4, and BA.5 lineages. In silico validation of the NTD156-7 and NTD25-7 primers and probes compared with publicly available SARS-CoV-2 genome databases showed low variability in regions corresponding to oligonucleotide binding sites. Similarly, in vitro validation with NGS-confirmed samples showed excellent correlation. RT-qPCR assays allow for near-real-time monitoring of circulating and emerging variants allowing for ongoing surveillance of variant dynamics in a local population. By performing periodic sequencing of variant surveillance by RT-qPCR methods, we were able to provide ongoing validation of the results obtained by RT-qPCR screening. Rapid SARS-CoV-2 variant identification and surveillance by this combined approach served to inform clinical decisions in a timely manner and permitted better utilization of sequencing resources.
Keywords: COVID-19; RT-qPCR; SARS-CoV-2; VOC; delta; surveillance.
Conflict of interest statement
All authors declare no conflict of interest.
Figures

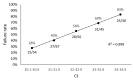

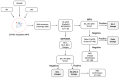
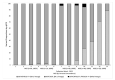
References
-
- WHO World Health Organization (WHO) Tracking SARS-CoV-2 Variants. [(accessed on 26 February 2021)]. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
-
- CDC SARS-CoV-2 Variant Classifications and Definitions. [(accessed on 26 February 2021)]; Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classificatio....
-
- ECDC SARS-CoV-2 Variants of Concern as of 24 February 2022. [(accessed on 26 February 2022)]. Available online: https://www.ecdc.europa.eu/en/covid-19/variants-concern.
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

