Influence of Canonical and Non-Canonical IFNLR1 Isoform Expression on Interferon Lambda Signaling
- PMID: 36992341
- PMCID: PMC10052089
- DOI: 10.3390/v15030632
Influence of Canonical and Non-Canonical IFNLR1 Isoform Expression on Interferon Lambda Signaling
Abstract
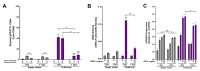
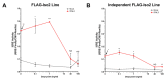
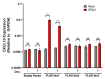
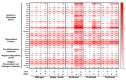
Interferon lambdas (IFNLs) are innate immune cytokines that induce antiviral cellular responses by signaling through a heterodimer composed of IL10RB and the interferon lambda receptor 1 (IFNLR1). Multiple IFNLR1 transcriptional variants are expressed in vivo and are predicted to encode distinct protein isoforms whose function is not fully established. IFNLR1 isoform 1 has the highest relative transcriptional expression and encodes the full-length functional form that supports canonical IFNL signaling. IFNLR1 isoforms 2 and 3 have lower relative expression and are predicted to encode signaling-defective proteins. To gain insight into IFNLR1 function and regulation, we explored how altering relative expression of IFNLR1 isoforms influenced the cellular response to IFNLs. To achieve this, we generated and functionally characterized stable HEK293T clones expressing doxycycline-inducible FLAG-tagged IFNLR1 isoforms. Minimal FLAG-IFNLR1 isoform 1 overexpression markedly increased IFNL3-dependent expression of antiviral and pro-inflammatory genes, a phenotype that could not be further augmented by expressing higher levels of FLAG-IFNLR1 isoform 1. Expression of low levels of FLAG-IFNLR1 isoform 2 led to partial induction of antiviral genes, but not pro-inflammatory genes, after IFNL3 treatment, a phenotype that was largely abrogated at higher FLAG-IFNLR1 isoform 2 expression levels. Expression of FLAG-IFNLR1 isoform 3 partially augmented antiviral gene expression after IFNL3 treatment. In addition, FLAG-IFNLR1 isoform 1 significantly reduced cellular sensitivity to the type-I IFN IFNA2 when overexpressed. These results identify a unique influence of canonical and non-canonical IFNLR1 isoforms on mediating the cellular response to interferons and provide insight into possible pathway regulation in vivo.
Keywords: antiviral; inflammation; innate immunity; interferon lambda receptor 1; interferons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

