Human Genome Polymorphisms and Computational Intelligence Approach Revealed a Complex Genomic Signature for COVID-19 Severity in Brazilian Patients
- PMID: 36992353
- PMCID: PMC10059592
- DOI: 10.3390/v15030645
Human Genome Polymorphisms and Computational Intelligence Approach Revealed a Complex Genomic Signature for COVID-19 Severity in Brazilian Patients
Abstract
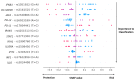
We present a genome polymorphisms/machine learning approach for severe COVID-19 prognosis. Ninety-six Brazilian severe COVID-19 patients and controls were genotyped for 296 innate immunity loci. Our model used a feature selection algorithm, namely recursive feature elimination coupled with a support vector machine, to find the optimal loci classification subset, followed by a support vector machine with the linear kernel (SVM-LK) to classify patients into the severe COVID-19 group. The best features that were selected by the SVM-RFE method included 12 SNPs in 12 genes: PD-L1, PD-L2, IL10RA, JAK2, STAT1, IFIT1, IFIH1, DC-SIGNR, IFNB1, IRAK4, IRF1, and IL10. During the COVID-19 prognosis step by SVM-LK, the metrics were: 85% accuracy, 80% sensitivity, and 90% specificity. In comparison, univariate analysis under the 12 selected SNPs showed some highlights for individual variant alleles that represented risk (PD-L1 and IFIT1) or protection (JAK2 and IFIH1). Variant genotypes carrying risk effects were represented by PD-L2 and IFIT1 genes. The proposed complex classification method can be used to identify individuals who are at a high risk of developing severe COVID-19 outcomes even in uninfected conditions, which is a disruptive concept in COVID-19 prognosis. Our results suggest that the genetic context is an important factor in the development of severe COVID-19.
Keywords: COVID-19 genetics; SARS-CoV-2 infection; complex genomic classifier; machine learning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Johns Hopkins University COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU) [(accessed on 22 January 2023)]. Available online: https://coronavirus.jhu.edu/map.html.
-
- Buitrago-Garcia D., Egli-Gany D., Counotte M.J., Hossmann S., Imeri H., Ipekci A.M., Salanti G., Low N. Occurrence and Transmission Potential of Asymptomatic and Presymptomatic SARSCoV-2 Infections: A Living Systematic Review and Meta-Analysis. PLoS Med. 2020;17:e1003346. doi: 10.1371/journal.pmed.1003346. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

