Genome Characterisation of the CGMMV Virus Population in Australia-Informing Plant Biosecurity Policy
- PMID: 36992452
- PMCID: PMC10051534
- DOI: 10.3390/v15030743
Genome Characterisation of the CGMMV Virus Population in Australia-Informing Plant Biosecurity Policy
Abstract
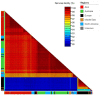
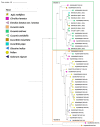
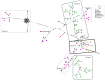
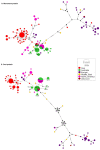
The detection of cucumber green mottle mosaic (CGMMV) in the Northern Territory (NT), Australia, in 2014 led to the introduction of strict quarantine measures for the importation of cucurbit seeds by the Australian federal government. Further detections in Queensland, Western Australia (WA), New South Wales and South Australia occurred in the period 2015-2020. To explore the diversity of the current Australian CGMMV population, 35 new coding sequence complete genomes for CGMMV isolates from Australian incursions and surveys were prepared for this study. In conjunction with published genomes from the NT and WA, sequence, phylogenetic, and genetic variation and variant analyses were performed, and the data were compared with those for international CGMMV isolates. Based on these analyses, it can be inferred that the Australian CGMMV population resulted from a single virus source via multiple introductions.
Keywords: cucumber green mottle mosaic virus; genetic diversity; median-joining network.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Hollings M., Komuro Y., Tochihara H. DPV Cucumber Green Mottle Mosaic Virus. [(accessed on 5 May 2021)]. Available online: https://www.dpvweb.net/dpv/showdpv/?dpvno=154.
-
- Ainsworth G.C. Mosaic Diseases of the Cucumber. Ann. Appl. Biol. 1935;22:55–67. doi: 10.1111/j.1744-7348.1935.tb07708.x. - DOI
-
- Boubourakas I.N., Hatziloukas E., Antignus Y., Katis N.I. Etiology of Leaf Chlorosis and Deterioration of the Fruit Interior of Watermelon Plants. J. Phytopathol. 2004;152:580–588. doi: 10.1111/j.1439-0434.2004.00900.x. - DOI
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

