Circulating Plasma Exosomal Proteins of Either SHIV-Infected Rhesus Macaque or HIV-Infected Patient Indicates a Link to Neuropathogenesis
- PMID: 36992502
- PMCID: PMC10058833
- DOI: 10.3390/v15030794
Circulating Plasma Exosomal Proteins of Either SHIV-Infected Rhesus Macaque or HIV-Infected Patient Indicates a Link to Neuropathogenesis
Abstract
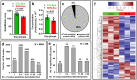
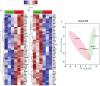
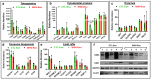
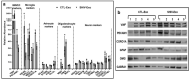
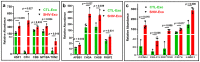
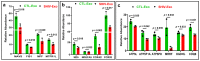
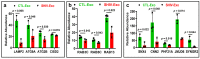
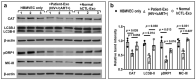
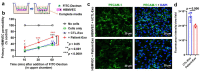
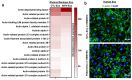
Despite the suppression of human immunodeficiency virus (HIV) replication by combined antiretroviral therapy (cART), 50-60% of HIV-infected patients suffer from HIV-associated neurocognitive disorders (HAND). Studies are uncovering the role of extracellular vesicles (EVs), especially exosomes, in the central nervous system (CNS) due to HIV infection. We investigated links among circulating plasma exosomal (crExo) proteins and neuropathogenesis in simian/human immunodeficiency virus (SHIV)-infected rhesus macaques (RM) and HIV-infected and cART treated patients (Patient-Exo). Isolated EVs from SHIV-infected (SHIV-Exo) and uninfected (CTL-Exo) RM were predominantly exosomes (particle size < 150 nm). Proteomic analysis quantified 5654 proteins, of which 236 proteins (~4%) were significantly, differentially expressed (DE) between SHIV-/CTL-Exo. Interestingly, different CNS cell specific markers were abundantly expressed in crExo. Proteins involved in latent viral reactivation, neuroinflammation, neuropathology-associated interactive as well as signaling molecules were expressed at significantly higher levels in SHIV-Exo than CTL-Exo. However, proteins involved in mitochondrial biogenesis, ATP production, autophagy, endocytosis, exocytosis, and cytoskeleton organization were significantly less expressed in SHIV-Exo than CTL-Exo. Interestingly, proteins involved in oxidative stress, mitochondrial biogenesis, ATP production, and autophagy were significantly downregulated in primary human brain microvascular endothelial cells exposed with HIV+/cART+ Patient-Exo. We showed that Patient-Exo significantly increased blood-brain barrier permeability, possibly due to loss of platelet endothelial cell adhesion molecule-1 protein and actin cytoskeleton structure. Our novel findings suggest that circulating exosomal proteins expressed CNS cell markers-possibly associated with viral reactivation and neuropathogenesis-that may elucidate the etiology of HAND.
Keywords: HIV-1; SHIV; circulating plasma exosomes; neuropathogenesis; proteomic analysis; rhesus macaque.
Conflict of interest statement
The authors declare no competing interest with respect to the research, authorship, and/or publication of this research article.
Figures
Similar articles
-
Novel Transmitted/Founder Simian-Human Immunodeficiency Viruses for Human Immunodeficiency Virus Latency and Cure Research.J Virol. 2020 Mar 31;94(8):e01659-19. doi: 10.1128/JVI.01659-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31969435 Free PMC article.
-
Central Nervous System Inflammation and Infection during Early, Nonaccelerated Simian-Human Immunodeficiency Virus Infection in Rhesus Macaques.J Virol. 2018 May 14;92(11):e00222-18. doi: 10.1128/JVI.00222-18. Print 2018 Jun 1. J Virol. 2018. PMID: 29563297 Free PMC article.
-
Simian-Human Immunodeficiency Virus SHIV.C.CH505 Persistence in ART-Suppressed Infant Macaques Is Characterized by Elevated SHIV RNA in the Gut and a High Abundance of Intact SHIV DNA in Naive CD4+ T Cells.J Virol. 2020 Dec 22;95(2):e01669-20. doi: 10.1128/JVI.01669-20. Print 2020 Dec 22. J Virol. 2020. PMID: 33087463 Free PMC article.
-
HIV-Proteins-Associated CNS Neurotoxicity, Their Mediators, and Alternative Treatments.Cell Mol Neurobiol. 2022 Nov;42(8):2553-2569. doi: 10.1007/s10571-021-01151-x. Epub 2021 Sep 25. Cell Mol Neurobiol. 2022. PMID: 34562223 Free PMC article. Review.
-
Neurological impact of HIV/AIDS and substance use alters brain function and structure.Front Med (Lausanne). 2025 Jan 7;11:1505440. doi: 10.3389/fmed.2024.1505440. eCollection 2024. Front Med (Lausanne). 2025. PMID: 39839621 Free PMC article. Review.
Cited by
-
Untargeted Metabolomic Profiling of Extracellular Vesicles Isolated from Human Seminal Plasma.Biomolecules. 2024 Sep 26;14(10):1211. doi: 10.3390/biom14101211. Biomolecules. 2024. PMID: 39456146 Free PMC article.
-
Bridging brain and blood: a prospective view on neuroimaging-exosome correlations in HIV-associated neurocognitive disorders.Front Neurol. 2025 Jan 7;15:1479272. doi: 10.3389/fneur.2024.1479272. eCollection 2024. Front Neurol. 2025. PMID: 39839878 Free PMC article. Review.
-
Fibrinogen in mice cerebral microvessels induces blood-brain barrier dysregulation with aging via a dynamin-related protein 1-dependent pathway.Geroscience. 2024 Feb;46(1):395-415. doi: 10.1007/s11357-023-00988-y. Epub 2023 Oct 28. Geroscience. 2024. PMID: 37897653 Free PMC article.
-
CROI 2023: Neuropsychiatric Complications in People With HIV.Top Antivir Med. 2023 Jun 30;31(4):543-555. Top Antivir Med. 2023. PMID: 37704201 Free PMC article. Review.
References
-
- UNAIDS Global HIV Statistics—Fact Sheet 2021. 2021. [(accessed on 14 March 2021)]. Available online: https://www.unaids.org/sites/default/files/media_asset/UNAIDS_FactSheet_....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

