This is a preprint.
Primate TRIM34 is a broadly-acting, TRIM5-dependent lentiviral restriction factor
- PMID: 36993223
- PMCID: PMC10055373
- DOI: 10.1101/2023.03.24.534139
Primate TRIM34 is a broadly-acting, TRIM5-dependent lentiviral restriction factor
Update in
-
Primate TRIM34 is a broadly-acting, TRIM5-dependent lentiviral restriction factor.Retrovirology. 2023 Aug 22;20(1):15. doi: 10.1186/s12977-023-00629-4. Retrovirology. 2023. PMID: 37608289 Free PMC article.
Abstract
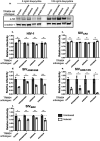
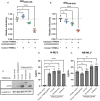
Human immunodeficiency virus (HIV) and other lentiviruses adapt to new hosts by evolving to evade host-specific innate immune proteins that differ in sequence and often viral recognition between host species. Understanding how these host antiviral proteins, called restriction factors, constrain lentivirus replication and transmission is key to understanding the emergence of pandemic viruses like HIV-1. Human TRIM34, a paralogue of the well-characterized lentiviral restriction factor TRIM5α, was previously identified by our lab via CRISPR-Cas9 screening as a restriction factor of certain HIV and SIV capsids. Here, we show that diverse primate TRIM34 orthologues from non-human primates can restrict a range of Simian Immunodeficiency Virus (SIV) capsids including SIV AGM-SAB , SIV AGM-TAN and SIV MAC capsids, which infect sabaeus monkeys, tantalus monkeys, and rhesus macaques, respectively. All primate TRIM34 orthologues tested, regardless of species of origin, were able to restrict this same subset of viral capsids. However, in all cases, this restriction also required the presence of TRIM5α. We demonstrate that TRIM5α is necessary, but not sufficient, for restriction of these capsids, and that human TRIM5α functionally interacts with TRIM34 from different species. Finally, we find that both the TRIM5α SPRY v1 loop and the TRIM34 SPRY domain are essential for TRIM34-mediated restriction. These data support a model in which TRIM34 is a broadly-conserved primate lentiviral restriction factor that acts in tandem with TRIM5α, such that together, these proteins can restrict capsids that neither can restrict alone.
Figures


References
-
- Nisole S, Stoye JP, Saïb A. TRIM family proteins: Retroviral restriction and antiviral defence. Nat Rev Microbiol. Nature Publishing Group; 2005;3:799–808. - PubMed
-
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–53. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
