This is a preprint.
Inhibitory input directs astrocyte morphogenesis through glial GABA B R
- PMID: 36993256
- PMCID: PMC10054985
- DOI: 10.1101/2023.03.14.532493
Inhibitory input directs astrocyte morphogenesis through glial GABA B R
Update in
-
Inhibitory input directs astrocyte morphogenesis through glial GABABR.Nature. 2023 May;617(7960):369-376. doi: 10.1038/s41586-023-06010-x. Epub 2023 Apr 26. Nature. 2023. PMID: 37100909 Free PMC article.
Abstract
Communication between neurons and glia plays an important role in establishing and maintaining higher order brain function. Astrocytes are endowed with complex morphologies which places their peripheral processes in close proximity to neuronal synapses and directly contributes to their regulation of brain circuits. Recent studies have shown that excitatory neuronal activity promotes oligodendrocyte differentiation; whether inhibitory neurotransmission regulates astrocyte morphogenesis during development is unknown. Here we show that inhibitory neuron activity is necessary and sufficient for astrocyte morphogenesis. We found that input from inhibitory neurons functions through astrocytic GABA B R and that its deletion in astrocytes results in a loss of morphological complexity across a host of brain regions and disruption of circuit function. Expression of GABA B R in developing astrocytes is regulated in a region-specific manner by SOX9 or NFIA and deletion of these transcription factors results in region-specific defects in astrocyte morphogenesis, which is conferred by interactions with transcription factors exhibiting region-restricted patterns of expression. Together our studies identify input from inhibitory neurons and astrocytic GABA B R as universal regulators of morphogenesis, while further revealing a combinatorial code of region-specific transcriptional dependencies for astrocyte development that is intertwined with activity-dependent processes.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
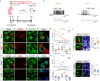
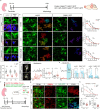
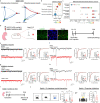
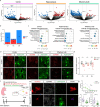


References
-
- Allen N. J. Astrocyte Regulation of Synaptic Behavior. Annu. Rev. Cell Dev. Biol. 30, 439–463 (2014). - PubMed
-
- Khakh B. S. & Deneen B. The Emerging Nature of Astrocyte Diversity. Annu Rev Neurosci 42, 187–207 (2019). - PubMed
-
- Volterra A. & Meldolesi J. Astrocytes, from brain glue to communication elements: the revolution continues. Nat Rev Neurosci 6, 626–640 (2005). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
