This is a preprint.
Genetic regulation of fetal hemoglobin across global populations
- PMID: 36993312
- PMCID: PMC10055601
- DOI: 10.1101/2023.03.24.23287659
Genetic regulation of fetal hemoglobin across global populations
Abstract
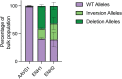
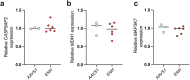
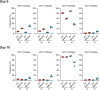
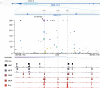
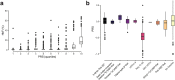
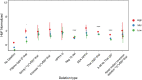
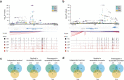
Human genetic variation has enabled the identification of several key regulators of fetal-to-adult hemoglobin switching, including BCL11A, resulting in therapeutic advances. However, despite the progress made, limited further insights have been obtained to provide a fuller accounting of how genetic variation contributes to the global mechanisms of fetal hemoglobin (HbF) gene regulation. Here, we have conducted a multi-ancestry genome-wide association study of 28,279 individuals from several cohorts spanning 5 continents to define the architecture of human genetic variation impacting HbF. We have identified a total of 178 conditionally independent genome-wide significant or suggestive variants across 14 genomic windows. Importantly, these new data enable us to better define the mechanisms by which HbF switching occurs in vivo. We conduct targeted perturbations to define BACH2 as a new genetically-nominated regulator of hemoglobin switching. We define putative causal variants and underlying mechanisms at the well-studied BCL11A and HBS1L-MYB loci, illuminating the complex variant-driven regulation present at these loci. We additionally show how rare large-effect deletions in the HBB locus can interact with polygenic variation to influence HbF levels. Our study paves the way for the next generation of therapies to more effectively induce HbF in sickle cell disease and β-thalassemia.
Figures







References
-
- Menzel S, Garner C, Gut I, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet. 2007;39(10):1197–1199. - PubMed
-
- Sankaran VG, Menne TF, Xu J, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322(5909):1839–1842. - PubMed
Publication types
Grants and funding
- U2C OD023196/OD/NIH HHS/United States
- OT2 OD026551/OD/NIH HHS/United States
- U24 OD023121/OD/NIH HHS/United States
- OT2 OD026552/OD/NIH HHS/United States
- OT2 OD025337/OD/NIH HHS/United States
- OT2 OD026550/OD/NIH HHS/United States
- OT2 OD026553/OD/NIH HHS/United States
- R01 HL068959/HL/NHLBI NIH HHS/United States
- OT2 OD025276/OD/NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- U24 OD023163/OD/NIH HHS/United States
- OT2 OD026556/OD/NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- U24 OD023176/OD/NIH HHS/United States
- OT2 OD026548/OD/NIH HHS/United States
- U01 HL120393/HL/NHLBI NIH HHS/United States
- OT2 OD025315/OD/NIH HHS/United States
- OT2 OD026549/OD/NIH HHS/United States
- MR/L003120/1/MRC_/Medical Research Council/United Kingdom
- OT2 OD025277/OD/NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- HHSN268201500015C/HL/NHLBI NIH HHS/United States
- HHSN268201600033C/ES/NIEHS NIH HHS/United States
- OT2 OD026555/OD/NIH HHS/United States
- OT2 OD023205/OD/NIH HHS/United States
- R01 HL146500/HL/NHLBI NIH HHS/United States
- OT2 OD026557/OD/NIH HHS/United States
- R01 DK103794/DK/NIDDK NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- OT2 OD026554/OD/NIH HHS/United States
- R01 HL079915/HL/NHLBI NIH HHS/United States
- OT2 OD023206/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
