This is a preprint.
DnaJC7 specifically regulates tau seeding
- PMID: 36993367
- PMCID: PMC10055123
- DOI: 10.1101/2023.03.16.532880
DnaJC7 specifically regulates tau seeding
Update in
-
DnaJC7 specifically regulates tau seeding.Elife. 2023 Jun 30;12:e86936. doi: 10.7554/eLife.86936. Elife. 2023. PMID: 37387473 Free PMC article.
Abstract
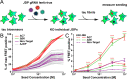
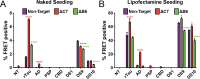
Neurodegenerative tauopathies are caused by accumulation of toxic tau protein assemblies. This appears to involve template-based seeding events, whereby tau monomer changes conformation and is recruited to a growing aggregate. Several large families of chaperone proteins, including Hsp70s and J domain proteins (JDPs) cooperate to regulate the folding of intracellular proteins such as tau, but the factors that coordinate this activity are not well known. The JDP DnaJC7 binds tau and reduces its intracellular aggregation. However, it is unknown whether this is specific to DnaJC7 or if other JDPs might be similarly involved. We used proteomics within a cell model to determine that DnaJC7 co-purified with insoluble tau and colocalized with intracellular aggregates. We individually knocked out every possible JDP and tested the effect on intracellular aggregation and seeding. DnaJC7 knockout decreased aggregate clearance and increased intracellular tau seeding. This depended on the ability of the J domain (JD) of DnaJC7 to bind to Hsp70, as JD mutations that block binding to Hsp70 abrogated the protective activity. Disease-associated mutations in the JD and substrate binding site of DnaJC7 also abrogated its protective activity. DnaJC7 thus specifically regulates tau aggregation in cooperation with Hsp70.
Conflict of interest statement
Competing Interests
The authors declare that they have no competing interests.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
