This is a preprint.
A Spatiotemporal Map of Co-Receptor Signaling Networks Underlying B Cell Activation
- PMID: 36993395
- PMCID: PMC10055206
- DOI: 10.1101/2023.03.17.533227
A Spatiotemporal Map of Co-Receptor Signaling Networks Underlying B Cell Activation
Update in
-
A spatiotemporal map of co-receptor signaling networks underlying B cell activation.Cell Rep. 2024 Jun 25;43(6):114332. doi: 10.1016/j.celrep.2024.114332. Epub 2024 Jun 7. Cell Rep. 2024. PMID: 38850533 Free PMC article.
Abstract
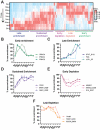
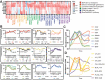
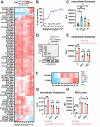
The B cell receptor (BCR) signals together with a multi-component co-receptor complex to initiate B cell activation in response to antigen binding. This process underlies nearly every aspect of proper B cell function. Here, we take advantage of peroxidase-catalyzed proximity labeling combined with quantitative mass spectrometry to track B cell co-receptor signaling dynamics from 10 seconds to 2 hours after BCR stimulation. This approach enables tracking of 2,814 proximity-labeled proteins and 1,394 quantified phosphosites and provides an unbiased and quantitative molecular map of proteins recruited to the vicinity of CD19, the key signaling subunit of the co-receptor complex. We detail the recruitment kinetics of essential signaling effectors to CD19 following activation, and then identify new mediators of B cell activation. In particular, we show that the glutamate transporter SLC1A1 is responsible for mediating rapid metabolic reprogramming immediately downstream of BCR stimulation and for maintaining redox homeostasis during B cell activation. This study provides a comprehensive map of the BCR signaling pathway and a rich resource for uncovering the complex signaling networks that regulate B cell activation.
Figures


References
-
- Kristyanto H., Blomberg N.J., Slot L.M., van der Voort E.I.H., Kerkman P.F., Bakker A., Burgers L.E., ten Brinck R.M., van der Helm-van Mil A.H.M., Spits H., et al. (2020). Persistently activated, proliferative memory autoreactive B cells promote inflammation in rheumatoid arthritis. Sci. Transl. Med. 12, eaaz5327. 10.1126/scitranslmed.aaz5327. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
