This is a preprint.
Non-additive effects of schizophrenia risk genes reflect convergent downstream function
- PMID: 36993466
- PMCID: PMC10055596
- DOI: 10.1101/2023.03.20.23287497
Non-additive effects of schizophrenia risk genes reflect convergent downstream function
Abstract
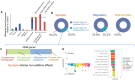
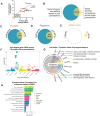
Genetic studies of schizophrenia (SCZ) reveal a complex polygenic risk architecture comprised of hundreds of risk variants, the majority of which are common in the population at-large and confer only modest increases in disorder risk. Precisely how genetic variants with individually small predicted effects on gene expression combine to yield substantial clinical impacts in aggregate is unclear. Towards this, we previously reported that the combinatorial perturbation of four SCZ risk genes ("eGenes", whose expression is regulated by common variants) resulted in gene expression changes that were not predicted by individual perturbations, being most non-additive among genes associated with synaptic function and SCZ risk. Now, across fifteen SCZ eGenes, we demonstrate that non-additive effects are greatest within groups of functionally similar eGenes. Individual eGene perturbations reveal common downstream transcriptomic effects ("convergence"), while combinatorial eGene perturbations result in changes that are smaller than predicted by summing individual eGene effects ("sub-additive effects"). Unexpectedly, these convergent and sub-additive downstream transcriptomic effects overlap and constitute a large proportion of the genome-wide polygenic risk score, suggesting that functional redundancy of eGenes may be a major mechanism underlying non-additivity. Single eGene perturbations likewise fail to predict the magnitude or directionality of cellular phenotypes resulting from combinatorial perturbations. Overall, our results indicate that polygenic risk cannot be extrapolated from experiments testing one risk gene at a time and must instead be empirically measured. By unravelling the interactions between complex risk variants, it may be possible to improve the clinical utility of polygenic risk scores through more powerful prediction of symptom onset, clinical trajectory, and treatment response, or to identify novel targets for therapeutic intervention.
Keywords: CRISPR; human induced pluripotent stem cells; neurons; psychiatric genomics; schizophrenia.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT E.S. is today an employee at Regeneron.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
