This is a preprint.
Automated evaluation of cardiac contractile dynamics and aging prediction using machine learning in a Drosophila model
- PMID: 36993511
- PMCID: PMC10055502
- DOI: 10.21203/rs.3.rs-2635745/v1
Automated evaluation of cardiac contractile dynamics and aging prediction using machine learning in a Drosophila model
Update in
-
Automated assessment of cardiac dynamics in aging and dilated cardiomyopathy Drosophila models using machine learning.Commun Biol. 2024 Jun 7;7(1):702. doi: 10.1038/s42003-024-06371-7. Commun Biol. 2024. PMID: 38849449 Free PMC article.
Abstract
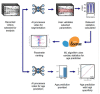
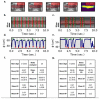
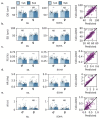
The Drosophila model has proven tremendously powerful for understanding pathophysiological bases of several human disorders including aging and cardiovascular disease. Relevant high-speed imaging and high-throughput lab assays generate large volumes of high-resolution videos, necessitating next-generation methods for rapid analysis. We present a platform for deep learning-assisted segmentation applied to optical microscopy of Drosophila hearts and the first to quantify cardiac physiological parameters during aging. An experimental test dataset is used to validate a Drosophila aging model. We then use two novel methods to predict fly aging: deep-learning video classification and machine-learning classification via cardiac parameters. Both models suggest excellent performance, with an accuracy of 83.3% (AUC 0.90) and 77.1% (AUC 0.85), respectively. Furthermore, we report beat-level dynamics for predicting the prevalence of cardiac arrhythmia. The presented approaches can expedite future cardiac assays for modeling human diseases in Drosophila and can be extended to numerous animal/human cardiac assays under multiple conditions.
Keywords: Drosophila heart model; Machine learning; aging prediction; cardiovascular disease; medical segmentation.
Conflict of interest statement
Additional Declarations: There is NO Competing Interest. Competing interests: none.
Figures



References
-
- Jean-Louis G., Kripke D. F., Ancoli-Israel S., Klauber M. R. & Sepulveda R. S. Sleep duration, illumination, and activity patterns in a population sample: effects of gender and ethnicity. Biol. Psychiatry 47, 921–927 (2000). - PubMed
-
- Bodmer R. & Venkatesh T. V. Heart development in Drosophila and vertebrates: Conservation of molecular mechanisms. Dev. Genet. 22, 181–186 (1998). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

