This is a preprint.
Neurodesk: An accessible, flexible, and portable data analysis environment for reproducible neuroimaging
- PMID: 36993557
- PMCID: PMC10055538
- DOI: 10.21203/rs.3.rs-2649734/v1
Neurodesk: An accessible, flexible, and portable data analysis environment for reproducible neuroimaging
Update in
-
Neurodesk: an accessible, flexible and portable data analysis environment for reproducible neuroimaging.Nat Methods. 2024 May;21(5):804-808. doi: 10.1038/s41592-023-02145-x. Epub 2024 Jan 8. Nat Methods. 2024. PMID: 38191935 Free PMC article.
Abstract
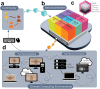
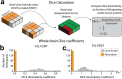
Neuroimaging data analysis often requires purpose-built software, which can be challenging to install and may produce different results across computing environments. Beyond being a roadblock to neuroscientists, these issues of accessibility and portability can hamper the reproducibility of neuroimaging data analysis pipelines. Here, we introduce the Neurodesk platform, which harnesses software containers to support a comprehensive and growing suite of neuroimaging software (https://www.neurodesk.org/). Neurodesk includes a browser-accessible virtual desktop environment and a command line interface, mediating access to containerized neuroimaging software libraries on various computing platforms, including personal and high-performance computers, cloud computing and Jupyter Notebooks. This community-oriented, open-source platform enables a paradigm shift for neuroimaging data analysis, allowing for accessible, flexible, fully reproducible, and portable data analysis pipelines.
Conflict of interest statement
Competing interests The authors declare no financial conflicts of interest.
Figures


References
-
- Brand A., Allen L., Altman M., Hlava M. & Scott J. Beyond authorship: attribution, contribution, collaboration, and credit. Learn. Publ. 28, 151–155 (2015).
-
- The FAIR Guiding Principles for scientific data management and stewardship ∣ Scientific Data. https://www.nature.com/articles/sdata201618. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

