This is a preprint.
A Molecular, Spatial, and Regulatory Atlas of the Hydra vulgaris Nervous System
- PMID: 36993575
- PMCID: PMC10055148
- DOI: 10.1101/2023.03.15.531610
A Molecular, Spatial, and Regulatory Atlas of the Hydra vulgaris Nervous System
Update in
-
A molecular, spatial and regulatory atlas of the Hydra vulgaris nervous system.Development. 2025 Oct 15;152(20):dev204983. doi: 10.1242/dev.204983. Epub 2025 Oct 24. Development. 2025. PMID: 41071669
Abstract
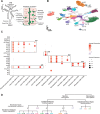
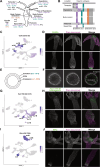
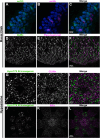
Hydra vulgaris, a cnidarian with a simple nerve net, is an emerging model for developmental, regenerative, and functional neuroscience. Its genetic tractability and capacity for whole-system imaging make it well suited for studying neuron replacement, regeneration, and neural circuit function. Here, we present the most comprehensive molecular and spatial characterization of the H. vulgaris nervous system to date. Using single-cell RNA sequencing, we identified eight neuron types, each defined by distinct neuropeptide expression, and further resolved these into fifteen transcriptionally distinct subtypes with unique spatial distributions and morphologies. To investigate the gene regulatory networks underlying neuronal differentiation, we applied trajectory inference, identified key transcription factors, and performed ATAC-seq on sorted neurons to map chromatin accessibility. All datasets are available through an interactive, user-friendly web portal to support broad use by the research community. Together, these resources provide a foundation for uncovering molecular mechanisms that govern nervous system development, homeostasis, and regeneration in H. vulgaris.
Conflict of interest statement
Competing Interests No competing interests declared.
Figures




References
-
- Badhiwala K. N., Gonzales D. L., Vercosa D. G., Avants B. W. and Robinson J. T. (2018). Microfluidics for electrophysiology, imaging, and behavioral analysis of Hydra. Lab Chip 18, 2523–2539. - PubMed
-
- Bode H., Berking S., David C. N., Gierer A., Schaller H. and Trenkner E. (1973). Quantitative analysis of cell types during growth and morphogenesis in Hydra. Wilhelm Roux Arch Entwickl Mech Org 171, 269–285. - PubMed
-
- Bode H. R., Heimfeld S., Koizumi O., Littlefield C. L. and Yaross M. S. (1988). Maintenance and Regeneration of the Nerve Net in Hydra. American Zoologist 28, 1053–1063.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
