This is a preprint.
Single cell and spatial alternative splicing analysis with long read sequencing
- PMID: 36993612
- PMCID: PMC10055662
- DOI: 10.21203/rs.3.rs-2674892/v1
Single cell and spatial alternative splicing analysis with long read sequencing
Update in
-
Single cell and spatial alternative splicing analysis with Nanopore long read sequencing.Nat Commun. 2025 Jul 19;16(1):6654. doi: 10.1038/s41467-025-60902-2. Nat Commun. 2025. PMID: 40683866 Free PMC article.
Abstract
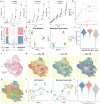
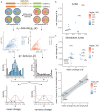
Long-read sequencing has become a powerful tool for alternative splicing analysis. However, technical and computational challenges have limited our ability to explore alternative splicing at single cell and spatial resolution. The higher sequencing error of long reads, especially high indel rates, have limited the accuracy of cell barcode and unique molecular identifier (UMI) recovery. Read truncation and mapping errors, the latter exacerbated by the higher sequencing error rates, can cause the false detection of spurious new isoforms. Downstream, there is yet no rigorous statistical framework to quantify splicing variation within and between cells/spots. In light of these challenges, we developed Longcell, a statistical framework and computational pipeline for accurate isoform quantification for single cell and spatial spot barcoded long read sequencing data. Longcell performs computationally efficient cell/spot barcode extraction, UMI recovery, and UMI-based truncation- and mapping-error correction. Through a statistical model that accounts for varying read coverage across cells/spots, Longcell rigorously quantifies the level of inter-cell/spot versus intra-cell/ spot diversity in exon-usage and detects changes in splicing distributions between cell populations. Applying Longcell to single cell long-read data from multiple contexts, we found that intra-cell splicing heterogeneity, where multiple isoforms co-exist within the same cell, is ubiquitous for highly expressed genes. On matched single cell and Visium long read sequencing for a tissue of colorectal cancer metastasis to the liver, Longcell found concordant signals between the two data modalities. Finally, on a perturbation experiment for 9 splicing factors, Longcell identified regulatory targets that are validated by targeted sequencing.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

