This is a preprint.
Beyond the reference: gene expression variation and transcriptional response to RNAi in C. elegans
- PMID: 36993640
- PMCID: PMC10055391
- DOI: 10.1101/2023.03.24.533964
Beyond the reference: gene expression variation and transcriptional response to RNAi in C. elegans
Update in
-
Beyond the reference: gene expression variation and transcriptional response to RNA interference in Caenorhabditis elegans.G3 (Bethesda). 2023 Aug 9;13(8):jkad112. doi: 10.1093/g3journal/jkad112. G3 (Bethesda). 2023. PMID: 37221008 Free PMC article.
Abstract
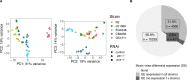
A universal feature of living systems is that natural variation in genotype underpins variation in phenotype. Yet, research in model organisms is often constrained to a single genetic background, the reference strain. Further, genomic studies that do evaluate wild strains typically rely on the reference strain genome for read alignment, leading to the possibility of biased inferences based on incomplete or inaccurate mapping; the extent of reference bias can be difficult to quantify. As an intermediary between genome and organismal traits, gene expression is well positioned to describe natural variability across genotypes generally and in the context of environmental responses, which can represent complex adaptive phenotypes. C. elegans sits at the forefront of investigation into small-RNA gene regulatory mechanisms, or RNA interference (RNAi), and wild strains exhibit natural variation in RNAi competency following environmental triggers. Here, we examine how genetic differences among five wild strains affect the C. elegans transcriptome in general and after inducing RNAi responses to two germline target genes. Approximately 34% of genes were differentially expressed across strains; 411 genes were not expressed at all in at least one strain despite robust expression in others, including 49 genes not expressed in reference strain N2. Despite the presence of hyper-diverse hotspots throughout the C. elegans genome, reference mapping bias was of limited concern: over 92% of variably expressed genes were robust to mapping issues. Overall, the transcriptional response to RNAi was strongly strain-specific and highly specific to the target gene, and the laboratory strain N2 was not representative of the other strains. Moreover, the transcriptional response to RNAi was not correlated with RNAi phenotypic penetrance; the two germline RNAi incompetent strains exhibited substantial differential gene expression following RNAi treatment, indicating an RNAi response despite failure to reduce expression of the target gene. We conclude that gene expression, both generally and in response to RNAi, differs across C. elegans strains such that choice of strain may meaningfully influence scientific conclusions. To provide a public, easily accessible resource for querying gene expression variation in this dataset, we introduce an interactive website at https://wildworm.biosci.gatech.edu/rnai/ .
Figures


References
-
- Ahringer J. (2006). Reverse genetics. In Ambros V. (Ed.), WormBook. 10.1895/wormbook.1.47.1 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
