This is a preprint.
The Genetic and Epigenetic Features of Bilateral Wilms Tumor Predisposition: A Report from the Children's Oncology Group AREN18B5-Q Study
- PMID: 36993649
- PMCID: PMC10055651
- DOI: 10.21203/rs.3.rs-2675436/v1
The Genetic and Epigenetic Features of Bilateral Wilms Tumor Predisposition: A Report from the Children's Oncology Group AREN18B5-Q Study
Update in
-
Genetic and epigenetic features of bilateral Wilms tumor predisposition in patients from the Children's Oncology Group AREN18B5-Q.Nat Commun. 2023 Dec 18;14(1):8006. doi: 10.1038/s41467-023-43730-0. Nat Commun. 2023. PMID: 38110397 Free PMC article.
Abstract
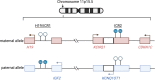
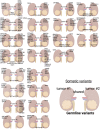
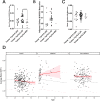
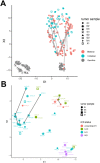
This study comprehensively evaluated the landscape of genetic and epigenetic events that predispose to synchronous bilateral Wilms tumor (BWT). We performed whole exome or whole genome sequencing, total-strand RNA-seq, and DNA methylation analysis using germline and/or tumor samples from 68 patients with BWT from St. Jude Children's Research Hospital and the Children's Oncology Group. We found that 25/61 (41%) of patients evaluated harbored pathogenic or likely pathogenic germline variants, with WT1 (14.8%), NYNRIN (6.6%), TRIM28 (5%) and the BRCA-related genes (5%) BRCA1, BRCA2, and PALB2 being most common. Germline WT1 variants were strongly associated with somatic paternal uniparental disomy encompassing the 11p15.5 and 11p13/WT1 loci and subsequent acquired pathogenic CTNNB1 variants. Somatic coding variants or genome-wide copy number alterations were almost never shared between paired synchronous BWT, suggesting that the acquisition of independent somatic variants leads to tumor formation in the context of germline or early embryonic, post-zygotic initiating events. In contrast, 11p15.5 status (loss of heterozygosity, loss or retention of imprinting) was shared among paired synchronous BWT in all but one case. The predominant molecular events for BWT predisposition include pathogenic germline variants or post-zygotic epigenetic hypermethylation at the 11p15.5 H19/ICR1 locus (loss of imprinting). This study demonstrates that post-zygotic somatic mosaicism for 11p15.5 hypermethylation/loss of imprinting is the single most common initiating molecular event predisposing to BWT. Evidence of somatic mosaicism for 11p15.5 loss of imprinting was detected in leukocytes of a cohort of BWT patients and long-term survivors, but not in unilateral Wilms tumor patients and long-term survivors or controls, further supporting the hypothesis that post-zygotic 11p15.5 alterations occurred in the mesoderm of patients who go on to develop BWT. Due to the preponderance of BWT patients with demonstrable germline or early embryonic tumor predisposition, BWT exhibits a unique biology when compared to unilateral Wilms tumor and therefore warrants continued refinement of its own treatment-relevant biomarkers which in turn may inform directed treatment strategies in the future.
Keywords: 11p15.5; Bilateral Wilms tumor; WT1; Wilms tumor predisposition.
Conflict of interest statement
Conflict of Interest Statement: The authors have no conflicts of interest to declare.
Figures




References
-
- Knudson A.G. & Strong L.C. Mutation and cancer: a model for Wilms’ tumor of the kidney. J Natl Cancer Inst 48, 313–24 (1972). - PubMed
-
- Spreafico F. et al. Wilms tumour. Nat Rev Dis Primers 7, 75 (2021). - PubMed
-
- Beckwith J.B., Kiviat N.B. & Bonadio J.F. Nephrogenic rests, nephroblastomatosis, and the pathogenesis of Wilms’ tumor. Pediatr Pathol 10, 1–36 (1990). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

