Synthesis, molecular docking, and binding Gibbs free energy calculation of β-nitrostyrene derivatives: Potential inhibitors of SARS-CoV-2 3CL protease
- PMID: 36993878
- PMCID: PMC10033154
- DOI: 10.1016/j.molstruc.2023.135409
Synthesis, molecular docking, and binding Gibbs free energy calculation of β-nitrostyrene derivatives: Potential inhibitors of SARS-CoV-2 3CL protease
Abstract
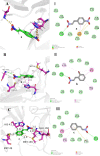
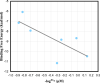
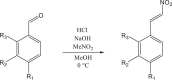
The outbreak of novel coronavirus disease 2019 (COVID-19), caused by the novel coronavirus (SARS-CoV-2), has had a significant impact on human health and the economic development. SARS-CoV-2 3CL protease (3CLpro) is highly conserved and plays a key role in mediating the transcription of virus replication. It is an ideal target for the design and screening of anti-coronavirus drugs. In this work, seven β-nitrostyrene derivatives were synthesized by Henry reaction and β-dehydration reaction, and their inhibitory effects on SARS-CoV-2 3CL protease were identified by enzyme activity inhibition assay in vitro. Among them, 4-nitro-β-nitrostyrene (compound a) showed the lowest IC50 values of 0.7297 µM. To investigate the key groups that determine the activity of β-nitrostyrene derivatives and their interaction mode with the receptor, the molecular docking using the CDOCKER protocol in Discovery Studio 2016 was performed. The results showed that the hydrogen bonds between β-NO2 and receptor GLY-143 and the π-π stacking between the aryl ring of the ligand and the imidazole ring of receptor HIS-41 significantly contributed to the ligand activity. Furthermore, the ligand-receptor absolute binding Gibbs free energies were calculated using the Binding Affinity Tool (BAT.py) to verify its correlation with the activity of β-nitrostyrene 3CLpro inhibitors as a scoring function. The higher correlation(r2=0.6) indicates that the absolute binding Gibbs free energy based on molecular dynamics can be used to predict the activity of new β-nitrostyrene 3CLpro inhibitors. These results provide valuable insights for the functional group-based design, structure optimization and the discovery of high accuracy activity prediction means of anti-COVID-19 lead compounds.
Keywords: 3CLpro inhibitor; Binding Gibbs free energies; Molecular docking; Receptor-ligand interaction; SARS-CoV-2; β-nitrostyrene derivatives.
© 2023 Published by Elsevier B.V.
Conflict of interest statement
All co-authors declare that there are no conflicts of interest.
Figures



Similar articles
-
Identification of Darunavir Derivatives for Inhibition of SARS-CoV-2 3CLpro.Int J Mol Sci. 2022 Dec 16;23(24):16011. doi: 10.3390/ijms232416011. Int J Mol Sci. 2022. PMID: 36555652 Free PMC article.
-
In silico analysis and identification of antiviral coumarin derivatives against 3-chymotrypsin-like main protease of the novel coronavirus SARS-CoV-2.Mol Divers. 2022 Apr;26(2):1053-1076. doi: 10.1007/s11030-021-10230-6. Epub 2021 Jul 2. Mol Divers. 2022. PMID: 34213728 Free PMC article.
-
Identification of Potential Inhibitors of 3CL Protease of SARS-CoV-2 From ZINC Database by Molecular Docking-Based Virtual Screening.Front Mol Biosci. 2020 Dec 17;7:603037. doi: 10.3389/fmolb.2020.603037. eCollection 2020. Front Mol Biosci. 2020. PMID: 33392261 Free PMC article.
-
An Integrated Computational and Experimental Approach to Identifying Inhibitors for SARS-CoV-2 3CL Protease.Front Mol Biosci. 2021 May 17;8:661424. doi: 10.3389/fmolb.2021.661424. eCollection 2021. Front Mol Biosci. 2021. PMID: 34079818 Free PMC article.
-
Biflavonoid as potential 3-chymotrypsin-like protease (3CLpro) inhibitor of SARS-Coronavirus.Results Chem. 2021 Jan;3:100087. doi: 10.1016/j.rechem.2020.100087. Epub 2020 Dec 25. Results Chem. 2021. PMID: 33520632 Free PMC article. Review.
Cited by
-
Design, synthesis and biological activity of peptidyl β-nitrostyrenes as cysteine protease inhibitors against Leishmania donovani.RSC Adv. 2025 Feb 19;15(8):5703-5719. doi: 10.1039/d4ra06510g. eCollection 2025 Feb 19. RSC Adv. 2025. PMID: 39981004 Free PMC article.
-
Isovaleryl Sucrose Esters from Atractylodes japonica and Their Cytotoxic Activity.Molecules. 2024 Jun 27;29(13):3069. doi: 10.3390/molecules29133069. Molecules. 2024. PMID: 38999021 Free PMC article.
-
Design, synthesis, and biological activity evaluation of dihydromyricetin derivatives against SARS-CoV-2-Omicron virus.J Enzyme Inhib Med Chem. 2024 Dec;39(1):2390909. doi: 10.1080/14756366.2024.2390909. Epub 2024 Aug 29. J Enzyme Inhib Med Chem. 2024. PMID: 39206852 Free PMC article.
-
Binding Mechanism of Inhibitors to BRD4 and BRD9 Decoded by Multiple Independent Molecular Dynamics Simulations and Deep Learning.Molecules. 2024 Apr 19;29(8):1857. doi: 10.3390/molecules29081857. Molecules. 2024. PMID: 38675678 Free PMC article.
References
-
- Health World. Organization COVID-19 vaccine tracker. and landscape. 2022
-
- Hashimoto M., Nagata N., Homma T., Maeda H., Dohi K., Seki N.M., Yoshihara K., Iwata-Yoshikawa N., Shiwa-Sudo N., Sakai Y., Shirakura M., Kishida N., Arita T., Suzuki Y., Watanabe S., Asanuma H., Sonoyama T., Suzuki T., Omoto S., Hasegawa H. Immunogenicity and protective efficacy of SARS-CoV-2 recombinant S-protein vaccine S-268019-b in cynomolgus monkeys. Vaccine. 2022;40(31):4231–4241. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

