A degradome-based prognostic signature that correlates with immune infiltration and tumor mutation burden in breast cancer
- PMID: 36993976
- PMCID: PMC10040797
- DOI: 10.3389/fimmu.2023.1140993
A degradome-based prognostic signature that correlates with immune infiltration and tumor mutation burden in breast cancer
Abstract
Introduction: Female breast cancer is the most common malignancy worldwide, with a high disease burden. The degradome is the most abundant class of cellular enzymes that play an essential role in regulating cellular activity. Dysregulation of the degradome may disrupt cellular homeostasis and trigger carcinogenesis. Thus we attempted to understand the prognostic role of degradome in breast cancer by means of establishing a prognostic signature based on degradome-related genes (DRGs) and assessed its clinical utility in multiple dimensions.
Methods: A total of 625 DRGs were obtained for analysis. Transcriptome data and clinical information of patients with breast cancer from TCGA-BRCA, METABRIC and GSE96058 were collected. NetworkAnalyst and cBioPortal were also utilized for analysis. LASSO regression analysis was employed to construct the degradome signature. Investigations of the degradome signature concerning clinical association, functional characterization, mutation landscape, immune infiltration, immune checkpoint expression and drug priority were orchestrated. Cell phenotype assays including colony formation, CCK8, transwell and wound healing were conducted in MCF-7 and MDA-MB-435S breast cancer cell lines, respectively.
Results: A 10-gene signature was developed and verified as an independent prognostic predictor combined with other clinicopathological parameters in breast cancer. The prognostic nomogram based on risk score (calculated based on the degradome signature) showed favourable capability in survival prediction and advantage in clinical benefit. High risk scores were associated with a higher degree of clinicopathological events (T4 stage and HER2-positive) and mutation frequency. Regulation of toll-like receptors and several cell cycle promoting activities were upregulated in the high-risk group. PIK3CA and TP53 mutations were dominant in the low- and high-risk groups, respectively. A significantly positive correlation was observed between the risk score and tumor mutation burden. The infiltration levels of immune cells and the expressions of immune checkpoints were significantly influenced by the risk score. Additionally, the degradome signature adequately predicted the survival of patients undergoing endocrinotherapy or radiotherapy. Patients in the low-risk group may achieve complete response after the first round of chemotherapy with cyclophosphamide and docetaxel, whereas patients in the high-risk group may benefit from 5-flfluorouracil. Several regulators of the PI3K/AKT/mTOR signaling pathway and the CDK family/PARP family were identified as potential molecular targets in the low- and high-risk groups, respectively. In vitro experiments further revealed that the knockdown of ABHD12 and USP41 significantly inhibit the proliferation, invasion and migration of breast cancer cells.
Conclusion: Multidimensional evaluation verified the clinical utility of the degradome signature in predicting prognosis, risk stratification and guiding treatment for patients with breast cancer.
Keywords: breast cancer; degradome; immune infiltration; immunotherapy; prognostic signature; tumour mutation burden.
Copyright © 2023 Luo, Ye, Chen, Zhang, Sun, Wang and Ou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





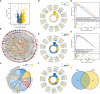
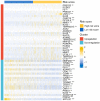


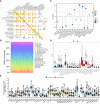

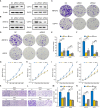
References
-
- Li W, Wang H, Dong S, Tang ZR, Chen L, Cai X, et al. Establishment and validation of a nomogram and web calculator for the risk of new vertebral compression fractures and cement leakage after percutaneous vertebroplasty in patients with osteoporotic vertebral compression fractures. Eur Spine J (2022) 31(5):1108–21. doi: 10.1007/s00586-021-07064-z - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

