Novel Human Meningioma Organoids Recapitulate the Aggressiveness of the Initiating Cell Subpopulations Identified by ScRNA-Seq
- PMID: 36994665
- PMCID: PMC10214266
- DOI: 10.1002/advs.202205525
Novel Human Meningioma Organoids Recapitulate the Aggressiveness of the Initiating Cell Subpopulations Identified by ScRNA-Seq
Abstract
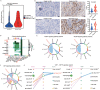
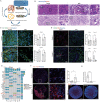
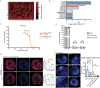
High-grade meningioma has an unsatisfactory outcome despite surgery and postoperative radiotherapy; however, the factors driving its malignancy and recurrence remain largely unknown, which limits the development of systemic treatments. Single-cell RNA sequencing (scRNA-Seq) technology is a powerful tool for studying intratumoral cellular heterogeneity and revealing the roles of various cell types in oncogenesis. In this study, scRNA-Seq is used to identify a unique initiating cell subpopulation (SULT1E1+ ) in high-grade meningiomas. This subpopulation modulates the polarization of M2-type macrophages and promotes meningioma progression and recurrence. A novel patient-derived meningioma organoid (MO) model is established to characterize this unique subpopulation. The resulting MOs fully retain the aggressiveness of SULT1E1+ and exhibit invasiveness in the brain after orthotopic transplantation. By targeting SULT1E1+ in MOs, the synthetic compound SRT1720 is identified as a potential agent for systemic treatment and radiation sensitization. These findings shed light on the mechanism underlying the malignancy of high-grade meningiomas and provide a novel therapeutic target for refractory high-grade meningioma.
Keywords: cell subpopulation; meningioma; patient-derived organoid model; single-cell RNA sequencing.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- a) Ostrom Q. T., Patil N., Cioffi G., Waite K., Kruchko C., Barnholtz‐Sloan J. S., Neuro Oncol. 2020, 22, iv1; - PMC - PubMed
- b) Huntoon K., Toland A. M. S., Dahiya S., Front. Oncol. 2020, 10, 579599; - PMC - PubMed
- c) Rogers L., Barani I., Chamberlain M., Kaley T. J., McDermott M., Raizer J., Schiff D., Weber D. C., Wen P. Y., Vogelbaum M. A., J Neurosurg Case Lessons 2015, 122, 4; - PMC - PubMed
- d) Lamszus K., J. Neuropathol. Exp. Neurol. 2004, 63, 275. - PubMed
-
- a) Goldbrunner R., Minniti G., Preusser M., Jenkinson M. D., Sallabanda K., Houdart E., von Deimling A., Stavrinou P., Lefranc F., Lund‐Johansen M., Moyal E. C.‐J., Brandsma D., Henriksson R., Soffietti R., Weller M., Lancet Oncol. 2016, 17, e383; - PubMed
- b) Apra C., Peyre M., Kalamarides M., Expert Rev. Neurother. 2018, 18, 241; - PubMed
- c) Riemenschneider M. J., Perry A., Reifenberger G., Lancet Neurol. 2006, 5, 1045. - PubMed
-
- Mawrin C., Perry A., J. Neuro‐Oncol. 2010, 99, 379. - PubMed
-
- Durand A., Labrousse F., Jouvet A., Bauchet L., Kalamarides M., Menei P., Deruty R., Moreau J. J., Fevre‐Montange M., Guyotat J., J. Neuro‐Oncol. 2009, 95, 367. - PubMed
-
- a) Birzu C., Peyre M., Sahm F., Curr Opin Oncol 2020, 32, 613; - PubMed
- b) Suppiah S., Nassiri F., Bi W. L., Dunn I. F., Hanemann C. O., Horbinski C. M., Hashizume R., James C. D., Mawrin C., Noushmehr H., Perry A., Sahm F., Sloan A., Von Deimling A., Wen P. Y., Aldape K., Zadeh G., International Consortium on M., Neuro Oncol 2019, 21, i4. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
