Unraveling the genetics of transformed splenic marginal zone lymphoma
- PMID: 36995085
- PMCID: PMC10368783
- DOI: 10.1182/bloodadvances.2022009415
Unraveling the genetics of transformed splenic marginal zone lymphoma
Abstract
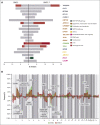
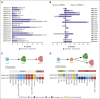
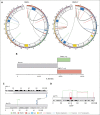
The genetic mechanisms associated with splenic marginal zone lymphoma (SMZL) transformation are not well defined. We studied 41 patients with SMZL that eventually underwent large B-cell lymphoma transformation. Tumor material was obtained either only at diagnosis (9 patients), at diagnosis and transformation (18 patients), and only at transformation (14 patients). Samples were categorized in 2 groups: (1) at diagnosis (SMZL, n = 27 samples), and (2) at transformation (SMZL-T, n = 32 samples). Using copy number arrays and a next-generation sequencing custom panel, we identified that the main genomic alterations in SMZL-T involved TNFAIP3, KMT2D, TP53, ARID1A, KLF2, 1q gains, and losses of 9p21.3 (CDKN2A/B) and 7q31-q32. Compared with SMZL, SMZL-T had higher genomic complexity, and higher incidence of TNFAIP3 and TP53 alterations, 9p21.3 (CDKN2A/B) losses, and 6p gains. SMZL and SMZL-T clones arose by divergent evolution from a common altered precursor cell that acquired different genetic alterations in virtually all evaluable cases (92%, 12 of 13 cases). Using whole-genome sequencing of diagnostic and transformation samples in 1 patient, we observed that the SMZL-T sample carried more genomic aberrations than the diagnostic sample, identified a translocation t(14;19)(q32;q13) present in both samples, and detected a focal B2M deletion due to chromothripsis acquired at transformation. Survival analysis showed that KLF2 mutations, complex karyotype, and International Prognostic Index score at transformation were predictive of a shorter survival from transformation (P = .001; P = .042; and P = .007; respectively). In summary, SMZL-T are characterized by higher genomic complexity than SMZL, and characteristic genomic alterations that could represent key players in the transformation event.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: F.N. has received honoraria from Janssen and AbbVie for speaking at educational activities. M.J.B. is currently an employee of Swedish Orphan Biovitrum. E.C. has been a consultant for Takeda, NanoString, AbbVie, and Illumina; has received research support from AstraZeneca; received honoraria from Janssen, EUSPharma, and Roche for speaking at educational activities; and is an inventor on a Lymphoma and Leukemia Molecular Profiling Project patent ‘Method for subtyping lymphoma subtypes by means of expression profiling’ (PCT/US2014/64161). A.L.-G. served on the advisory board of Roche, Celgene, Novartis, and Gilead/Kite, and received grants from Celgene and Gilead/Kite. The remaining authors declare no competing financial interests.
The current affiliation for M.J.B. is Swedish Orphan Biovitrum, Portugal.
Figures




References
-
- Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised. 4th ed. IARC; Lyon, France: 2017.
-
- Salido M, Baró C, Oscier D, et al. Cytogenetic aberrations and their prognostic value in a series of 330 splenic marginal zone B-cell lymphomas: a multicenter study of the Splenic B-Cell Lymphoma Group. Blood. 2010;116(9):1479–1488. - PubMed
-
- Jaramillo Oquendo C, Parker H, Oscier D, et al. The (epi)genomic landscape of splenic marginal zone lymphoma, biological implications, clinical utility, and future questions. J Transl Genet Genom. 2021;5(2):89–111.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

