Overexpression screen of chromosome 21 genes reveals modulators of Sonic hedgehog signaling relevant to Down syndrome
- PMID: 36995257
- PMCID: PMC10120076
- DOI: 10.1242/dmm.049712
Overexpression screen of chromosome 21 genes reveals modulators of Sonic hedgehog signaling relevant to Down syndrome
Abstract
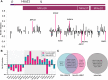
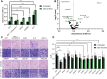
Trisomy 21 and mutations in the Sonic hedgehog (SHH) signaling pathway cause overlapping and pleiotropic phenotypes including cerebellar hypoplasia, craniofacial abnormalities, congenital heart defects and Hirschsprung disease. Trisomic cells derived from individuals with Down syndrome possess deficits in SHH signaling, suggesting that overexpression of human chromosome 21 genes may contribute to SHH-associated phenotypes by disrupting normal SHH signaling during development. However, chromosome 21 does not encode any known components of the canonical SHH pathway. Here, we sought to identify chromosome 21 genes that modulate SHH signaling by overexpressing 163 chromosome 21 cDNAs in a series of SHH-responsive mouse cell lines. We confirmed overexpression of trisomic candidate genes using RNA sequencing in the cerebella of Ts65Dn and TcMAC21 mice, model systems for Down syndrome. Our findings indicate that some human chromosome 21 genes, including DYRK1A, upregulate SHH signaling, whereas others, such as HMGN1, inhibit SHH signaling. Individual overexpression of four genes (B3GALT5, ETS2, HMGN1 and MIS18A) inhibits the SHH-dependent proliferation of primary granule cell precursors. Our study prioritizes dosage-sensitive chromosome 21 genes for future mechanistic studies. Identification of the genes that modulate SHH signaling may suggest new therapeutic avenues for ameliorating Down syndrome phenotypes.
Keywords: Aneuploidy; Down syndrome; Gene dosage effects; Genetic screen; Sonic hedgehog; Trisomy 21.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests M.O. is a CEO, employee and shareholder of Trans Chromosomics, Inc. Other authors declare no competing interests.
Figures




Similar articles
-
Chronic up-regulation of the SHH pathway normalizes some developmental effects of trisomy in Ts65Dn mice.Mech Dev. 2015 Feb;135:68-80. doi: 10.1016/j.mod.2014.11.004. Epub 2014 Dec 12. Mech Dev. 2015. PMID: 25511459 Free PMC article.
-
Forebrain Shh overexpression improves cognitive function and locomotor hyperactivity in an aneuploid mouse model of Down syndrome and its euploid littermates.Acta Neuropathol Commun. 2021 Aug 16;9(1):137. doi: 10.1186/s40478-021-01237-z. Acta Neuropathol Commun. 2021. PMID: 34399854 Free PMC article.
-
Chronic up-regulation of sonic hedgehog has little effect on postnatal craniofacial morphology of euploid and trisomic mice.Dev Dyn. 2016 Feb;245(2):114-22. doi: 10.1002/dvdy.24361. Epub 2015 Dec 6. Dev Dyn. 2016. PMID: 26509735 Free PMC article.
-
A Sonic hedgehog (Shh) response deficit in trisomic cells may be a common denominator for multiple features of Down syndrome.Prog Brain Res. 2012;197:223-36. doi: 10.1016/B978-0-444-54299-1.00011-X. Prog Brain Res. 2012. PMID: 22541295 Free PMC article. Review.
-
Down syndrome and mouse models.Curr Opin Genet Dev. 1998 Jun;8(3):316-21. doi: 10.1016/s0959-437x(98)80088-9. Curr Opin Genet Dev. 1998. PMID: 9690992 Review.
Cited by
-
Integrated analysis of immunometabolic interactions in Down syndrome.Sci Adv. 2024 Dec 13;10(50):eadq3073. doi: 10.1126/sciadv.adq3073. Epub 2024 Dec 13. Sci Adv. 2024. PMID: 39671500 Free PMC article.
-
Characterisation of a GNAS variant linked to cortisol-producing adrenocortical adenoma.Endocr Oncol. 2025 May 16;5(1):e250009. doi: 10.1530/EO-25-0009. eCollection 2025 Jan. Endocr Oncol. 2025. PMID: 40391091 Free PMC article.
-
Increased endothelial sclerostin caused by elevated DSCAM mediates multiple trisomy 21 phenotypes.J Clin Invest. 2024 Jun 3;134(11):e167811. doi: 10.1172/JCI167811. J Clin Invest. 2024. PMID: 38828726 Free PMC article.
-
Network Pharmacology Identifies Intersection Genes of Apigenin and Naringenin in Down Syndrome as Potential Therapeutic Targets.Pharmaceuticals (Basel). 2024 Aug 20;17(8):1090. doi: 10.3390/ph17081090. Pharmaceuticals (Basel). 2024. PMID: 39204195 Free PMC article.
-
Integration of ATAC-seq and RNA-seq identifies MX1-mediated AP-1 transcriptional regulation as a therapeutic target for Down syndrome.Biol Res. 2023 Dec 9;56(1):67. doi: 10.1186/s40659-023-00474-x. Biol Res. 2023. PMID: 38066591 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

