RNA Polymerase II transcription independent of TBP in murine embryonic stem cells
- PMID: 36995326
- PMCID: PMC10174690
- DOI: 10.7554/eLife.83810
RNA Polymerase II transcription independent of TBP in murine embryonic stem cells
Abstract
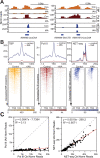
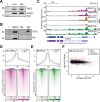
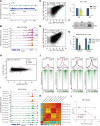
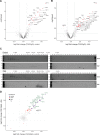
Transcription by RNA Polymerase II (Pol II) is initiated by the hierarchical assembly of the pre-initiation complex onto promoter DNA. Decades of research have shown that the TATA-box binding protein (TBP) is essential for Pol II loading and initiation. Here, we report instead that acute depletion of TBP in mouse embryonic stem cells has no global effect on ongoing Pol II transcription. In contrast, acute TBP depletion severely impairs RNA Polymerase III initiation. Furthermore, Pol II transcriptional induction occurs normally upon TBP depletion. This TBP-independent transcription mechanism is not due to a functional redundancy with the TBP paralog TRF2, though TRF2 also binds to promoters of transcribed genes. Rather, we show that the TFIID complex can form and, despite having reduced TAF4 and TFIIA binding when TBP is depleted, the Pol II machinery is sufficiently robust in sustaining TBP-independent transcription.
Keywords: chromosomes; degron; embryonic stem cells; gene expression; genomics; mouse; transcription.
© 2023, Kwan, Nguyen et al.
Conflict of interest statement
JK, TN, AU, MB, JC, JL, FV, PL, ST No competing interests declared
Figures














Update of
Comment in
- doi: 10.7554/eLife.88396
References
-
- Antonova SV, Haffke M, Corradini E, Mikuciunas M, Low TY, Signor L, van Es RM, Gupta K, Scheer E, Vos HR, Tora L, Heck AJR, Timmers HTM, Berger I. Chaperonin CCT checkpoint function in basal transcription factor TFIID assembly. Nature Structural & Molecular Biology. 2018;25:1119–1127. doi: 10.1038/s41594-018-0156-z. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

