Endothelial depletion of Atg7 triggers astrocyte-microvascular disassociation at blood-brain barrier
- PMID: 36995368
- PMCID: PMC10067974
- DOI: 10.1083/jcb.202103098
Endothelial depletion of Atg7 triggers astrocyte-microvascular disassociation at blood-brain barrier
Abstract
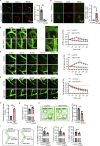
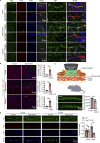
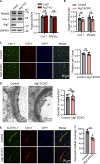
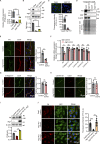
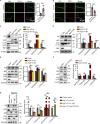
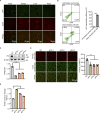
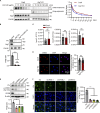
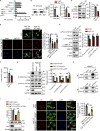
Microvascular basement membrane (BM) plays a pivotal role in the interactions of astrocyte with endothelium to maintain the blood-brain barrier (BBB) homeostasis; however, the significance and precise regulation of the endothelial cell-derived BM component in the BBB remain incompletely understood. Here, we report that conditional knockout of Atg7 in endothelial cells (Atg7-ECKO) leads to astrocyte-microvascular disassociation in the brain. Our results reveal astrocytic endfeet detachment from microvessels and BBB leakage in Atg7-ECKO mice. Furthermore, we find that the absence of endothelial Atg7 downregulates the expression of fibronectin, a major BM component of the BBB, causing significantly reduced coverage of astrocytes along cerebral microvessels. We reveal Atg7 triggers the expression of endothelial fibronectin via regulating PKA activity to affect the phosphorylation of cAMP-responsive element-binding protein. These results suggest that Atg7-regulated endothelial fibronectin production is required for astrocytes adhesion to microvascular wall for maintaining the BBB homeostasis. Thus, endothelial Atg7 plays an essential role in astrocyte-endothelium interactions to maintain the BBB integrity.
© 2023 Liu et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures





References
-
- Andreone, B.J., Chow B.W., Tata A., Lacoste B., Ben-Zvi A., Bullock K., Deik A.A., Ginty D.D., Clish C.B., and Gu C.. 2017. Blood-brain barrier permeability is regulated by lipid transport-dependent suppression of caveolae-mediated transcytosis. Neuron. 94:581–594.e5. 10.1016/j.neuron.2017.03.043 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

