Crystal Structure of an i-Motif from the HRAS Oncogene Promoter
- PMID: 36995904
- PMCID: PMC10330059
- DOI: 10.1002/anie.202301666
Crystal Structure of an i-Motif from the HRAS Oncogene Promoter
Abstract
An i-motif is a non-canonical DNA structure implicated in gene regulation and linked to cancers. The C-rich strand of the HRAS oncogene, 5'-CGCCCGTGCCCTGCGCCCGCAACCCGA-3' (herein referred to as iHRAS), forms an i-motif in vitro but its exact structure was unknown. HRAS is a member of the RAS proto-oncogene family. About 19 % of US cancer patients carry mutations in RAS genes. We solved the structure of iHRAS at 1.77 Å resolution. The structure reveals that iHRAS folds into a double hairpin. The two double hairpins associate in an antiparallel fashion, forming an i-motif dimer capped by two loops on each end and linked by a connecting region. Six C-C+ base pairs form each i-motif core, and the core regions are extended by a G-G base pair and a cytosine stacking. Extensive canonical and non-canonical base pairing and stacking stabilizes the connecting region and loops. The iHRAS structure is the first atomic resolution structure of an i-motif from a human oncogene. This structure sheds light on i-motifs folding and function in the cell.
Keywords: Dimers; Gene Regulation; Non-Canonical Base Pairing; X-Ray Crystallography; i-Motifs.
© 2023 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Figures


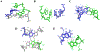

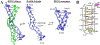
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

