Characterisation of the antiviral RNA interference response to Toscana virus in sand fly cells
- PMID: 36996243
- PMCID: PMC10112792
- DOI: 10.1371/journal.ppat.1011283
Characterisation of the antiviral RNA interference response to Toscana virus in sand fly cells
Abstract
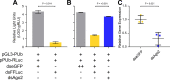
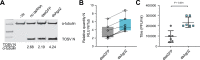
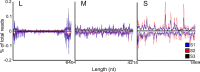
Toscana virus (TOSV) (Bunyavirales, Phenuiviridae, Phlebovirus, Toscana phlebovirus) and other related human pathogenic arboviruses are transmitted by phlebotomine sand flies. TOSV has been reported in nations bordering the Mediterranean Sea among other regions. Infection can result in febrile illness as well as meningitis and encephalitis. Understanding vector-arbovirus interactions is crucial to improving our knowledge of how arboviruses spread, and in this context, immune responses that control viral replication play a significant role. Extensive research has been conducted on mosquito vector immunity against arboviruses, with RNA interference (RNAi) and specifically the exogenous siRNA (exo-siRNA) pathway playing a critical role. However, the antiviral immunity of phlebotomine sand flies is less well understood. Here we were able to show that the exo-siRNA pathway is active in a Phlebotomus papatasi-derived cell line. Following TOSV infection, distinctive 21 nucleotide virus-derived small interfering RNAs (vsiRNAs) were detected. We also identified the exo-siRNA effector Ago2 in this cell line, and silencing its expression rendered the exo-siRNA pathway largely inactive. Thus, our data show that this pathway is active as an antiviral response against a sand fly transmitted bunyavirus, TOSV.
Copyright: © 2023 Alexander et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

