Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography
- PMID: 36996819
- PMCID: PMC7615348
- DOI: 10.1016/j.chom.2023.03.004
Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography
Abstract
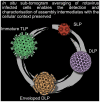
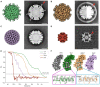
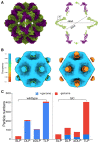
Rotavirus assembly is a complex process that involves the stepwise acquisition of protein layers in distinct intracellular locations to form the fully assembled particle. Understanding and visualization of the assembly process has been hampered by the inaccessibility of unstable intermediates. We characterize the assembly pathway of group A rotaviruses observed in situ within cryo-preserved infected cells through the use of cryoelectron tomography of cellular lamellae. Our findings demonstrate that the viral polymerase VP1 recruits viral genomes during particle assembly, as revealed by infecting with a conditionally lethal mutant. Additionally, pharmacological inhibition to arrest the transiently enveloped stage uncovered a unique conformation of the VP4 spike. Subtomogram averaging provided atomic models of four intermediate states, including a pre-packaging single-layered intermediate, the double-layered particle, the transiently enveloped double-layered particle, and the fully assembled triple-layered virus particle. In summary, these complementary approaches enable us to elucidate the discrete steps involved in forming an intracellular rotavirus particle.
Keywords: Reoviridae; cellular structural biology; cryoelectron tomography; dsRNA viruses; electron microscopy; in situ; rotavirus; subtomogram averaging; virus assembly; virus structure.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

